Clonal expansion of SIV-infected cells in macaques on antiretroviral therapy is similar to that of HIV-infected cells in humans
- PMID: 31291371
- PMCID: PMC6619828
- DOI: 10.1371/journal.ppat.1007869
Clonal expansion of SIV-infected cells in macaques on antiretroviral therapy is similar to that of HIV-infected cells in humans
Abstract
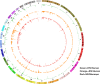
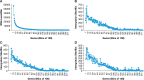
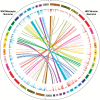
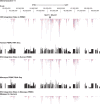
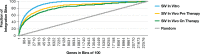
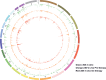
Clonal expansion of HIV infected cells plays an important role in the formation and persistence of the reservoir that allows the virus to persist, in DNA form, despite effective antiretroviral therapy. We used integration site analysis to ask if there is a similar clonal expansion of SIV infected cells in macaques. We show that the distribution of HIV and SIV integration sites in vitro is similar and that both viruses preferentially integrate in many of the same genes. We obtained approximately 8000 integration sites from blood samples taken from SIV-infected macaques prior to the initiation of ART, and from blood, spleen, and lymph node samples taken at necropsy. Seven clones were identified in the pre-ART samples; one persisted for a year on ART. An additional 100 clones were found only in on-ART samples; a number of these clones were found in more than one tissue. The timing and extent of clonal expansion of SIV-infected cells in macaques and HIV-infected cells in humans is quite similar. This suggests that SIV-infected macaques represent a useful model of the clonal expansion of HIV infected cells in humans that can be used to evaluate strategies intended to control or eradicate the viral reservoir.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Initiation of Antiretroviral Therapy Restores CD4+ T Memory Stem Cell Homeostasis in Simian Immunodeficiency Virus-Infected Macaques.J Virol. 2016 Jul 11;90(15):6699-6708. doi: 10.1128/JVI.00492-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27170752 Free PMC article.
-
Brain Macrophages in Simian Immunodeficiency Virus-Infected, Antiretroviral-Suppressed Macaques: a Functional Latent Reservoir.mBio. 2017 Aug 15;8(4):e01186-17. doi: 10.1128/mBio.01186-17. mBio. 2017. PMID: 28811349 Free PMC article.
-
Bone Marrow-Derived CD4+ T Cells Are Depleted in Simian Immunodeficiency Virus-Infected Macaques and Contribute to the Size of the Replication-Competent Reservoir.J Virol. 2018 Dec 10;93(1):e01344-18. doi: 10.1128/JVI.01344-18. Print 2019 Jan 1. J Virol. 2018. PMID: 30305357 Free PMC article.
-
Brain macrophages harbor latent, infectious simian immunodeficiency virus.AIDS. 2019 Dec 1;33 Suppl 2(Suppl 2):S181-S188. doi: 10.1097/QAD.0000000000002269. AIDS. 2019. PMID: 31789817 Free PMC article. Review.
-
HIV Persistence in Adipose Tissue Reservoirs.Curr HIV/AIDS Rep. 2018 Feb;15(1):60-71. doi: 10.1007/s11904-018-0378-z. Curr HIV/AIDS Rep. 2018. PMID: 29423731 Free PMC article. Review.
Cited by
-
Integration in oncogenes plays only a minor role in determining the in vivo distribution of HIV integration sites before or during suppressive antiretroviral therapy.PLoS Pathog. 2021 Apr 7;17(4):e1009141. doi: 10.1371/journal.ppat.1009141. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33826675 Free PMC article.
-
Early antiretroviral therapy in SIV-infected rhesus macaques reveals a multiphasic, saturable dynamic accumulation of the rebound competent viral reservoir.PLoS Pathog. 2024 Apr 9;20(4):e1012135. doi: 10.1371/journal.ppat.1012135. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38593120 Free PMC article.
-
The forces driving clonal expansion of the HIV-1 latent reservoir.Virol J. 2020 Jan 7;17(1):4. doi: 10.1186/s12985-019-1276-8. Virol J. 2020. PMID: 31910871 Free PMC article. Review.
-
SIV infection and ARV treatment reshape the transcriptional and epigenetic profile of naïve and memory T cells in vivo.J Virol. 2024 Jun 13;98(6):e0028324. doi: 10.1128/jvi.00283-24. Epub 2024 May 23. J Virol. 2024. PMID: 38780248 Free PMC article.
-
Clones of infected cells arise early in HIV-infected individuals.JCI Insight. 2019 Jun 20;4(12):e128432. doi: 10.1172/jci.insight.128432. eCollection 2019 Jun 20. JCI Insight. 2019. PMID: 31217357 Free PMC article.
References
-
- Kearney MF, Anderson EM, Coomer C, Smith L, Shao W, Johnson N, et al. Well-mixed plasma and tissue viral populations in RT-SHIV-infected macaques implies a lack of viral replication in the tissues during antiretroviral therapy. Retrovirology. 2015;12:93 10.1186/s12977-015-0212-2 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

