Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms
- PMID: 31289293
- PMCID: PMC6616478
- DOI: 10.1038/s41598-019-46143-6
Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms
Abstract
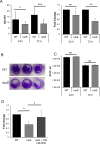
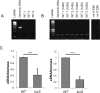
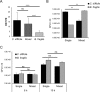
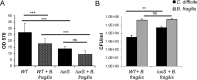
The anaerobic gut pathogen, Clostridioides difficile, forms adherent biofilms that may play an important role in recurrent C. difficile infections. The mechanisms underlying C. difficile community formation and inter-bacterial interactions are nevertheless poorly understood. C. difficile produces AI-2, a quorum sensing molecule that modulates biofilm formation across many bacterial species. We found that a strain defective in LuxS, the enzyme that mediates AI-2 production, is defective in biofilm development in vitro. Transcriptomic analyses of biofilms formed by wild type (WT) and luxS mutant (luxS) strains revealed a downregulation of prophage loci in the luxS mutant biofilms compared to the WT. Detection of phages and eDNA within biofilms may suggest that DNA release by phage-mediated cell lysis contributes to C. difficile biofilm formation. In order to understand if LuxS mediates C. difficile crosstalk with other gut species, C. difficile interactions with a common gut bacterium, Bacteroides fragilis, were studied. We demonstrate that C. difficile growth is significantly reduced when co-cultured with B. fragilis in mixed biofilms. Interestingly, the absence of C. difficile LuxS alleviates the B. fragilis-mediated growth inhibition. Dual species RNA-sequencing analyses from single and mixed biofilms revealed differential modulation of distinct metabolic pathways for C. difficile WT, luxS and B. fragilis upon co-culture, indicating that AI-2 may be involved in induction of selective metabolic responses in B. fragilis. Overall, our data suggest that C. difficile LuxS/AI-2 utilises different mechanisms to mediate formation of single and mixed species communities.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Streptococcus gordonii LuxS/autoinducer-2 quorum-sensing system modulates the dual-species biofilm formation with Streptococcus mutans.J Basic Microbiol. 2017 Jul;57(7):605-616. doi: 10.1002/jobm.201700010. Epub 2017 May 9. J Basic Microbiol. 2017. PMID: 28485524
-
Production of AI-2 is mediated by the S-ribosylhomocystein lyase gene luxS in Bacteroides fragilis and Bacteroides vulgatus.J Basic Microbiol. 2014 Jul;54(7):644-9. doi: 10.1002/jobm.201300311. Epub 2013 Sep 11. J Basic Microbiol. 2014. PMID: 24026770
-
The LuxS/AI-2 Quorum-Sensing System of Streptococcus pneumoniae Is Required to Cause Disease, and to Regulate Virulence- and Metabolism-Related Genes in a Rat Model of Middle Ear Infection.Front Cell Infect Microbiol. 2018 May 4;8:138. doi: 10.3389/fcimb.2018.00138. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29780750 Free PMC article.
-
The LuxS/AI-2 system of Streptococcus suis.Appl Microbiol Biotechnol. 2018 Sep;102(17):7231-7238. doi: 10.1007/s00253-018-9170-7. Epub 2018 Jun 25. Appl Microbiol Biotechnol. 2018. PMID: 29938319 Review.
-
The role of single and mixed biofilms in Clostridioides difficile infection and strategies for prevention and inhibition.Crit Rev Microbiol. 2024 May;50(3):285-299. doi: 10.1080/1040841X.2023.2189950. Epub 2023 Mar 20. Crit Rev Microbiol. 2024. PMID: 36939635 Review.
Cited by
-
Biofilm Formation of Clostridioides difficile, Toxin Production and Alternatives to Conventional Antibiotics in the Treatment of CDI.Microorganisms. 2023 Aug 26;11(9):2161. doi: 10.3390/microorganisms11092161. Microorganisms. 2023. PMID: 37764005 Free PMC article. Review.
-
Biofilm regulation in Clostridioides difficile: Novel systems linked to hypervirulence.PLoS Pathog. 2021 Sep 9;17(9):e1009817. doi: 10.1371/journal.ppat.1009817. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34499698 Free PMC article. Review.
-
Genetic diversity, biofilm formation, and Vancomycin resistance of clinical Clostridium innocuum isolates.BMC Microbiol. 2024 Sep 18;24(1):353. doi: 10.1186/s12866-024-03503-1. BMC Microbiol. 2024. PMID: 39294587 Free PMC article.
-
Challenges and opportunities of phage therapy for Klebsiella pneumoniae infections.Appl Environ Microbiol. 2024 Oct 23;90(10):e0135324. doi: 10.1128/aem.01353-24. Epub 2024 Sep 30. Appl Environ Microbiol. 2024. PMID: 39345202 Review.
-
The role of extracellular DNA in the formation, architecture, stability, and treatment of bacterial biofilms.Biotechnol Bioeng. 2021 Jun;118(6):2129-2141. doi: 10.1002/bit.27760. Epub 2021 Mar 27. Biotechnol Bioeng. 2021. PMID: 33748946 Free PMC article. Review.
References
-
- Davies KA, et al. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID) Lancet Infect Dis. 2014;14:1208–1219. doi: 10.1016/S1473-3099(14)70991-0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

