Positional Dynamics and Glycosomal Recruitment of Developmental Regulators during Trypanosome Differentiation
- PMID: 31289175
- PMCID: PMC6747725
- DOI: 10.1128/mBio.00875-19
Positional Dynamics and Glycosomal Recruitment of Developmental Regulators during Trypanosome Differentiation
Abstract
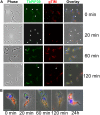
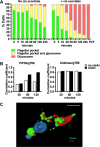
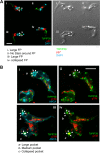
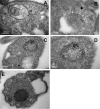
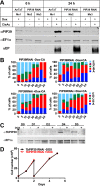
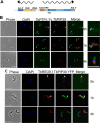
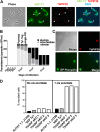
Glycosomes are peroxisome-related organelles that compartmentalize the glycolytic enzymes in kinetoplastid parasites. These organelles are developmentally regulated in their number and composition, allowing metabolic adaptation to the parasite's needs in the blood of mammalian hosts or within their arthropod vector. A protein phosphatase cascade regulates differentiation between parasite developmental forms, comprising a tyrosine phosphatase, Trypanosoma brucei PTP1 (TbPTP1), which dephosphorylates and inhibits a serine threonine phosphatase, TbPIP39, which promotes differentiation. When TbPTP1 is inactivated, TbPIP39 is activated and during differentiation becomes located in glycosomes. Here we have tracked TbPIP39 recruitment to glycosomes during differentiation from bloodstream "stumpy" forms to procyclic forms. Detailed microscopy and live-cell imaging during the synchronous transition between life cycle stages revealed that in stumpy forms, TbPIP39 is located at a periflagellar pocket site closely associated with TbVAP, which defines the flagellar pocket endoplasmic reticulum. TbPTP1 is also located at the same site in stumpy forms, as is REG9.1, a regulator of stumpy-enriched mRNAs. This site provides a molecular node for the interaction between TbPTP1 and TbPIP39. Within 30 min of the initiation of differentiation, TbPIP39 relocates to glycosomes, whereas TbPTP1 disperses to the cytosol. Overall, the study identifies a "stumpy regulatory nexus" (STuRN) that coordinates the molecular components of life cycle signaling and glycosomal development during transmission of Trypanosoma bruceiIMPORTANCE African trypanosomes are parasites of sub-Saharan Africa responsible for both human and animal disease. The parasites are transmitted by tsetse flies, and completion of their life cycle involves progression through several development steps. The initiation of differentiation between blood and tsetse fly forms is signaled by a phosphatase cascade, ultimately trafficked into peroxisome-related organelles called glycosomes that are unique to this group of organisms. Glycosomes undergo substantial remodeling of their composition and function during the differentiation step, but how this is regulated is not understood. Here we identify a cytological site where the signaling molecules controlling differentiation converge before the dispersal of one of them into glycosomes. In combination, the study provides the first insight into the spatial coordination of signaling pathway components in trypanosomes as they undergo cell-type differentiation.
Keywords: development; differentiation; glycosome; organelle; parasite; trypanosome.
Copyright © 2019 Szöőr et al.
Figures


Similar articles
-
A novel phosphatase cascade regulates differentiation in Trypanosoma brucei via a glycosomal signaling pathway.Genes Dev. 2010 Jun 15;24(12):1306-16. doi: 10.1101/gad.570310. Genes Dev. 2010. PMID: 20551176 Free PMC article.
-
Protein tyrosine phosphatase TbPTP1: A molecular switch controlling life cycle differentiation in trypanosomes.J Cell Biol. 2006 Oct 23;175(2):293-303. doi: 10.1083/jcb.200605090. Epub 2006 Oct 16. J Cell Biol. 2006. PMID: 17043136 Free PMC article.
-
Turnover of glycosomes during life-cycle differentiation of Trypanosoma brucei.Autophagy. 2008 Apr;4(3):294-308. doi: 10.4161/auto.5443. Autophagy. 2008. PMID: 18365344
-
Biogenesis, maintenance and dynamics of glycosomes in trypanosomatid parasites.Biochim Biophys Acta. 2016 May;1863(5):1038-48. doi: 10.1016/j.bbamcr.2015.09.015. Epub 2015 Sep 16. Biochim Biophys Acta. 2016. PMID: 26384872 Review.
-
Metabolic functions of glycosomes in trypanosomatids.Biochim Biophys Acta. 2006 Dec;1763(12):1463-77. doi: 10.1016/j.bbamcr.2006.08.019. Epub 2006 Aug 24. Biochim Biophys Acta. 2006. PMID: 17023066 Review.
Cited by
-
An Alba-domain protein required for proteome remodelling during trypanosome differentiation and host transition.PLoS Pathog. 2021 Jan 25;17(1):e1009239. doi: 10.1371/journal.ppat.1009239. eCollection 2021 Jan. PLoS Pathog. 2021. PMID: 33493187 Free PMC article.
-
Right place, right time: Environmental sensing and signal transduction directs cellular differentiation and motility in Trypanosoma brucei.Mol Microbiol. 2021 May;115(5):930-941. doi: 10.1111/mmi.14682. Mol Microbiol. 2021. PMID: 33434370 Free PMC article. Review.
-
Diverse Functions of Tim50, a Component of the Mitochondrial Inner Membrane Protein Translocase.Int J Mol Sci. 2021 Jul 21;22(15):7779. doi: 10.3390/ijms22157779. Int J Mol Sci. 2021. PMID: 34360547 Free PMC article. Review.
-
Structure, Properties, and Function of Glycosomes in Trypanosoma cruzi.Front Cell Infect Microbiol. 2020 Jan 31;10:25. doi: 10.3389/fcimb.2020.00025. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32083023 Free PMC article. Review.
-
Trypanosoma brucei Tim50 Possesses PAP Activity and Plays a Critical Role in Cell Cycle Regulation and Parasite Infectivity.mBio. 2021 Oct 26;12(5):e0159221. doi: 10.1128/mBio.01592-21. Epub 2021 Sep 14. mBio. 2021. PMID: 34517757 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

