Amino-acid inserts of HIV-1 capsid (CA) induce CA degradation and abrogate viral infectivity: Insights for the dynamics and mechanisms of HIV-1 CA decomposition
- PMID: 31285456
- PMCID: PMC6614453
- DOI: 10.1038/s41598-019-46082-2
Amino-acid inserts of HIV-1 capsid (CA) induce CA degradation and abrogate viral infectivity: Insights for the dynamics and mechanisms of HIV-1 CA decomposition
Abstract
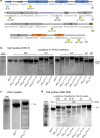
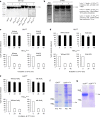
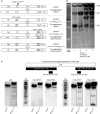
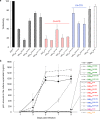
Accumulation of amino acid (AA) insertions/substitutions are observed in the Gag-protein of HIV-1 variants resistant to HIV-1 protease inhibitors. Here, we found that HIV-1 carrying AA insertions in capsid protein (CA) undergoes aberrant CA degradation. When we generated recombinant HIV-1s (rHIV-1s) containing 19-AAs in Gag, such insertions caused significant CA degradation, which initiated in CA's C-terminal. Such rHIV-1s had remarkable morphological abnormality, decreased infectivity, and no replicative ability, which correlated with levels of CA degradation. The CA degradation observed was energy-independent and had no association with cellular/viral proteolytic mechanisms, suggesting that the CA degradation occurs due to conformational/structural incompatibility caused by the 19-AA insertions. The incorporation of degradation-prone CA into the wild-type CA resulted in significant disruption of replication competence in "chimeric" virions. The data should allow better understanding of the dynamics and mechanisms of CA decomposition/degradation and retroviral uncoating, which may lead to new approach for antiretroviral modalities.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Allosteric Regulation of HIV-1 Capsid Structure for Gag Assembly, Virion Production, and Viral Infectivity by a Disordered Interdomain Linker.J Virol. 2019 Aug 13;93(17):e00381-19. doi: 10.1128/JVI.00381-19. Print 2019 Sep 1. J Virol. 2019. PMID: 31189701 Free PMC article.
-
Identification of a Structural Element in HIV-1 Gag Required for Virus Particle Assembly and Maturation.mBio. 2018 Oct 16;9(5):e01567-18. doi: 10.1128/mBio.01567-18. mBio. 2018. PMID: 30327442 Free PMC article.
-
HIV-1 Matrix Trimerization-Impaired Mutants Are Rescued by Matrix Substitutions That Enhance Envelope Glycoprotein Incorporation.J Virol. 2019 Dec 12;94(1):e01526-19. doi: 10.1128/JVI.01526-19. Print 2019 Dec 12. J Virol. 2019. PMID: 31619553 Free PMC article.
-
Human Immunodeficiency Virus Type 2 Capsid Protein Mutagenesis Reveals Amino Acid Residues Important for Virus Particle Assembly.J Mol Biol. 2022 Oct 15;434(19):167753. doi: 10.1016/j.jmb.2022.167753. Epub 2022 Jul 19. J Mol Biol. 2022. PMID: 35868362 Free PMC article.
-
Capsid (CA) protein as a novel drug target: recent progress in the research of HIV-1 CA inhibitors.Mini Rev Med Chem. 2009 Apr;9(4):510-8. doi: 10.2174/138955709787847958. Mini Rev Med Chem. 2009. PMID: 19356128 Review.
Cited by
-
A Small Molecule, ACAi-028, with Anti-HIV-1 Activity Targets a Novel Hydrophobic Pocket on HIV-1 Capsid.Antimicrob Agents Chemother. 2021 Sep 17;65(10):e0103921. doi: 10.1128/AAC.01039-21. Epub 2021 Jul 6. Antimicrob Agents Chemother. 2021. PMID: 34228546 Free PMC article.
-
Synthesis of Selenoesters via Aldol Condensation and/or Conjugate Reduction and Their Antiviral Activities.ACS Omega. 2022 Dec 27;8(1):1369-1374. doi: 10.1021/acsomega.2c06784. eCollection 2023 Jan 10. ACS Omega. 2022. PMID: 36643423 Free PMC article.
-
Label-free imaging of nuclear membrane for analysis of nuclear import of viral complexes.J Virol Methods. 2023 Dec;322:114834. doi: 10.1016/j.jviromet.2023.114834. Epub 2023 Oct 22. J Virol Methods. 2023. PMID: 37875225 Free PMC article.
-
Rotten to the core: antivirals targeting the HIV-1 capsid core.Retrovirology. 2021 Dec 22;18(1):41. doi: 10.1186/s12977-021-00583-z. Retrovirology. 2021. PMID: 34937567 Free PMC article. Review.
References
-
- Department of Health and Human Services. Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. (2106).
-
- Mitsuya, H. & Erickson, J. W. In Textbook Of AIDS Medicine. (eds Merigan, T. C., Bartlet, J. G. & Bolognes, D.), 751–780 (The Williams & Wilkins Co., Baltimore, Md., 1999).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

