Genome-wide functional association networks: background, data & state-of-the-art resources
- PMID: 31281921
- PMCID: PMC7373183
- DOI: 10.1093/bib/bbz064
Genome-wide functional association networks: background, data & state-of-the-art resources
Abstract
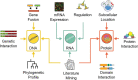
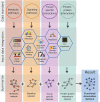
The vast amount of experimental data from recent advances in the field of high-throughput biology begs for integration into more complex data structures such as genome-wide functional association networks. Such networks have been used for elucidation of the interplay of intra-cellular molecules to make advances ranging from the basic science understanding of evolutionary processes to the more translational field of precision medicine. The allure of the field has resulted in rapid growth of the number of available network resources, each with unique attributes exploitable to answer different biological questions. Unfortunately, the high volume of network resources makes it impossible for the intended user to select an appropriate tool for their particular research question. The aim of this paper is to provide an overview of the underlying data and representative network resources as well as to mention methods of integration, allowing a customized approach to resource selection. Additionally, this report will provide a primer for researchers venturing into the field of network integration.
Keywords: Bayesian classification; functional association networks; network inference; protein–protein interactions.
© The Author(s) 2019. Published by Oxford University Press.
Figures
Similar articles
-
The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology: This report is based on a colloquium, “The Global Genome Question: Microbes as the Key to Understanding Evolution and Ecology,” sponsored by the American Academy of Microbiology and held October 11-13, 2002, in Longboat Key, Florida.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33119236 Free Books & Documents. Review.
-
Computational methods for Gene Regulatory Networks reconstruction and analysis: A review.Artif Intell Med. 2019 Apr;95:133-145. doi: 10.1016/j.artmed.2018.10.006. Epub 2018 Nov 9. Artif Intell Med. 2019. PMID: 30420244 Review.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
Mouse Genome Informatics (MGI): Resources for Mining Mouse Genetic, Genomic, and Biological Data in Support of Primary and Translational Research.Methods Mol Biol. 2017;1488:47-73. doi: 10.1007/978-1-4939-6427-7_3. Methods Mol Biol. 2017. PMID: 27933520
-
A Protocol for the Construction and Curation of Genome-Scale Integrated Metabolic and Regulatory Network Models.Methods Mol Biol. 2019;1927:203-214. doi: 10.1007/978-1-4939-9142-6_14. Methods Mol Biol. 2019. PMID: 30788794
Cited by
-
Network Crosstalk as a Basis for Drug Repurposing.Front Genet. 2022 Mar 8;13:792090. doi: 10.3389/fgene.2022.792090. eCollection 2022. Front Genet. 2022. PMID: 35350247 Free PMC article.
-
The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest.Nucleic Acids Res. 2023 Jan 6;51(D1):D638-D646. doi: 10.1093/nar/gkac1000. Nucleic Acids Res. 2023. PMID: 36370105 Free PMC article.
-
Selected cachaça yeast strains share a genomic profile related to traits relevant to industrial fermentation processes.Appl Environ Microbiol. 2024 Jan 24;90(1):e0175923. doi: 10.1128/aem.01759-23. Epub 2023 Dec 19. Appl Environ Microbiol. 2024. PMID: 38112453 Free PMC article.
-
Construction and contextualization approaches for protein-protein interaction networks.Comput Struct Biotechnol J. 2022 Jun 18;20:3280-3290. doi: 10.1016/j.csbj.2022.06.040. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832626 Free PMC article. Review.
-
Enhancing coevolutionary signals in protein-protein interaction prediction through clade-wise alignment integration.Sci Rep. 2024 Mar 12;14(1):6009. doi: 10.1038/s41598-024-55655-9. Sci Rep. 2024. PMID: 38472223 Free PMC article.

