Advances in cryo-electron tomography and subtomogram averaging and classification
- PMID: 31280905
- PMCID: PMC6863431
- DOI: 10.1016/j.sbi.2019.05.021
Advances in cryo-electron tomography and subtomogram averaging and classification
Abstract
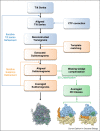
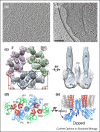
Cryo-electron tomography (cryoET) can provide 3D reconstructions, or tomograms, of pleomorphic objects such as organelles or cells in their close-to-native states. Subtomograms that contain repetitive structures can be further extracted and subjected to averaging and classification to improve resolution, and this process has become an emerging structural biology method referred to as cryoET subtomogram averaging and classification (cryoSTAC). Recent technical advances in cryoSTAC have had a profound impact on many fields in biology. Here, I review recent exciting work on several macromolecular assemblies demonstrating the power of cryoSTAC for in situ structure analysis and discuss challenges and future directions.
Copyright © 2019 The Author. Published by Elsevier Ltd.. All rights reserved.
Figures


Similar articles
-
Cryo-Electron Tomography and Subtomogram Averaging.Methods Enzymol. 2016;579:329-67. doi: 10.1016/bs.mie.2016.04.014. Epub 2016 Jun 22. Methods Enzymol. 2016. PMID: 27572733 Review.
-
Current data processing strategies for cryo-electron tomography and subtomogram averaging.Biochem J. 2021 May 28;478(10):1827-1845. doi: 10.1042/BCJ20200715. Biochem J. 2021. PMID: 34003255 Free PMC article. Review.
-
Toward high-resolution in situ structural biology with cryo-electron tomography and subtomogram averaging.Curr Opin Struct Biol. 2019 Oct;58:1-9. doi: 10.1016/j.sbi.2019.03.018. Epub 2019 Apr 18. Curr Opin Struct Biol. 2019. PMID: 31005754 Review.
-
Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION.Nat Protoc. 2016 Nov;11(11):2054-65. doi: 10.1038/nprot.2016.124. Epub 2016 Sep 29. Nat Protoc. 2016. PMID: 27685097 Free PMC article.
-
Subtomogram averaging from cryo-electron tomograms.Methods Cell Biol. 2019;152:217-259. doi: 10.1016/bs.mcb.2019.04.003. Epub 2019 May 15. Methods Cell Biol. 2019. PMID: 31326022 Review.
Cited by
-
Manipulation of the Cellular Membrane-Cytoskeleton Network for RNA Virus Replication and Movement in Plants.Viruses. 2023 Mar 14;15(3):744. doi: 10.3390/v15030744. Viruses. 2023. PMID: 36992453 Free PMC article. Review.
-
Single Cell Cryo-Soft X-ray Tomography Shows That Each Chlamydia Trachomatis Inclusion Is a Unique Community of Bacteria.Life (Basel). 2021 Aug 18;11(8):842. doi: 10.3390/life11080842. Life (Basel). 2021. PMID: 34440586 Free PMC article.
-
STOPGAP: an open-source package for template matching, subtomogram alignment and classification.Acta Crystallogr D Struct Biol. 2024 May 1;80(Pt 5):336-349. doi: 10.1107/S205979832400295X. Epub 2024 Apr 12. Acta Crystallogr D Struct Biol. 2024. PMID: 38606666 Free PMC article.
-
Structure and activity of particulate methane monooxygenase arrays in methanotrophs.Nat Commun. 2022 Sep 5;13(1):5221. doi: 10.1038/s41467-022-32752-9. Nat Commun. 2022. PMID: 36064719 Free PMC article.
-
Novel Secondary Ion Mass Spectrometry Methods for the Examination of Metabolic Effects at the Cellular and Subcellular Levels.Front Behav Neurosci. 2020 Jul 20;14:124. doi: 10.3389/fnbeh.2020.00124. eCollection 2020. Front Behav Neurosci. 2020. PMID: 32792922 Free PMC article.
References
-
- Henderson R. From electron crystallography to single particle cryoEM (nobel lecture) Angew Chem Int Ed Engl. 2018;57:10804–10825. - PubMed
-
- Koning R.I., Koster A.J., Sharp T.H. Advances in cryo-electron tomography for biology and medicine. Ann Anat. 2018;217:82–96. - PubMed
-
- Forster F., Hegerl R. Structure determination in situ by averaging of tomograms. Methods Cell Biol. 2007;79:741–767. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

