The Elusive 5'-Deoxyadenosyl Radical: Captured and Characterized by Electron Paramagnetic Resonance and Electron Nuclear Double Resonance Spectroscopies
- PMID: 31274303
- PMCID: PMC6784836
- DOI: 10.1021/jacs.9b05926
The Elusive 5'-Deoxyadenosyl Radical: Captured and Characterized by Electron Paramagnetic Resonance and Electron Nuclear Double Resonance Spectroscopies
Abstract
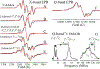
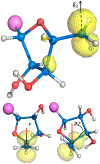
The 5'-deoxyadenosyl radical (5'-dAdo·) abstracts a substrate H atom as the first step in radical-based transformations catalyzed by adenosylcobalamin-dependent and radical S-adenosyl-l-methionine (RS) enzymes. Notwithstanding its central biological role, 5'-dAdo· has eluded characterization despite efforts spanning more than a half-century. Here, we report generation of 5'-dAdo· in a RS enzyme active site at 12 K using a novel approach involving cryogenic photoinduced electron transfer from the [4Fe-4S]+ cluster to the coordinated S-adenosylmethionine (SAM) to induce homolytic S-C5' bond cleavage. We unequivocally reveal the structure of this long-sought radical species through the use of electron paramagnetic resonance (EPR) and electron nuclear double resonance (ENDOR) spectroscopies with isotopic labeling, complemented by density-functional computations: a planar C5' (2pπ) radical (∼70% spin occupancy); the C5'(H)2 plane is rotated by ∼37° (experiment)/39° (DFT) relative to the C5'-C4'-(C4'-H) plane, placing a C5'-H antiperiplanar to the ribose-ring oxygen, which helps stabilize the radical against elimination of the 4'-H. The agreement between φ from experiment and in vacuo DFT indicates that the conformation is intrinsic to 5-dAdo· itself, and not determined by its environment.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
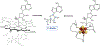
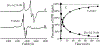
Similar articles
-
Mechanism of Radical Initiation in the Radical SAM Enzyme Superfamily.Annu Rev Biochem. 2023 Jun 20;92:333-349. doi: 10.1146/annurev-biochem-052621-090638. Epub 2023 Apr 4. Annu Rev Biochem. 2023. PMID: 37018846 Free PMC article. Review.
-
Paradigm Shift for Radical S-Adenosyl-l-methionine Reactions: The Organometallic Intermediate Ω Is Central to Catalysis.J Am Chem Soc. 2018 Jul 18;140(28):8634-8638. doi: 10.1021/jacs.8b04061. Epub 2018 Jul 6. J Am Chem Soc. 2018. PMID: 29954180 Free PMC article.
-
Radical SAM Enzyme Spore Photoproduct Lyase: Properties of the Ω Organometallic Intermediate and Identification of Stable Protein Radicals Formed during Substrate-Free Turnover.J Am Chem Soc. 2020 Oct 28;142(43):18652-18660. doi: 10.1021/jacs.0c08585. Epub 2020 Oct 15. J Am Chem Soc. 2020. PMID: 32966073 Free PMC article.
-
Why Nature Uses Radical SAM Enzymes so Widely: Electron Nuclear Double Resonance Studies of Lysine 2,3-Aminomutase Show the 5'-dAdo• "Free Radical" Is Never Free.J Am Chem Soc. 2015 Jun 10;137(22):7111-21. doi: 10.1021/jacs.5b00498. Epub 2015 May 19. J Am Chem Soc. 2015. PMID: 25923449 Free PMC article.
-
Radical SAM enzymes: Nature's choice for radical reactions.FEBS Lett. 2023 Jan;597(1):92-101. doi: 10.1002/1873-3468.14519. Epub 2022 Oct 27. FEBS Lett. 2023. PMID: 36251330 Free PMC article. Review.
Cited by
-
Radical S-adenosylmethionine maquette chemistry: Cx3Cx2C peptide coordinated redox active [4Fe-4S] clusters.J Biol Inorg Chem. 2019 Sep;24(6):793-807. doi: 10.1007/s00775-019-01708-8. Epub 2019 Sep 5. J Biol Inorg Chem. 2019. PMID: 31486952
-
Pyruvate formate-lyase activating enzyme: The catalytically active 5'-deoxyadenosyl radical caught in the act of H-atom abstraction.Proc Natl Acad Sci U S A. 2023 Nov 21;120(47):e2314696120. doi: 10.1073/pnas.2314696120. Epub 2023 Nov 13. Proc Natl Acad Sci U S A. 2023. PMID: 37956301 Free PMC article.
-
Mechanism of Radical Initiation in the Radical SAM Enzyme Superfamily.Annu Rev Biochem. 2023 Jun 20;92:333-349. doi: 10.1146/annurev-biochem-052621-090638. Epub 2023 Apr 4. Annu Rev Biochem. 2023. PMID: 37018846 Free PMC article. Review.
-
Iron-sulfur cluster-dependent enzymes and molybdenum-dependent reductases in the anaerobic metabolism of human gut microbes.Metallomics. 2024 Nov 7;16(11):mfae049. doi: 10.1093/mtomcs/mfae049. Metallomics. 2024. PMID: 39504489 Free PMC article. Review.
-
The B12-independent glycerol dehydratase activating enzyme from Clostridium butyricum cleaves SAM to produce 5'-deoxyadenosine and not 5'-deoxy-5'-(methylthio)adenosine.J Inorg Biochem. 2022 Feb;227:111662. doi: 10.1016/j.jinorgbio.2021.111662. Epub 2021 Nov 12. J Inorg Biochem. 2022. PMID: 34847521 Free PMC article.
References
-
- Lenhert PG; Hodgkin DC Structure of the 5,6-dimethylbenzimidazolylcobamide coenzyme. Nature 1961, 192, 937–938. - PubMed
-
- Halpern J Mechanisms of Coenzyme B12-Dependent Rearrangements. Science 1985, 227, 869–875. - PubMed
-
- Brown KL Chemistry and Enzymology of Vitamin B12. Chem. Rev 2005, 105, 2075–2149. - PubMed
-
- Banerjee R; Ragsdale SW The many faces of vitamine B12: Catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem 2003, 72, 209–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

