PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments
- PMID: 31270318
- PMCID: PMC6610634
- DOI: 10.1038/s41467-019-10755-3
PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments
Abstract
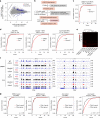
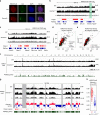
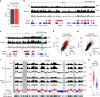
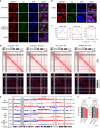
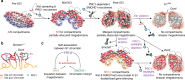
X-chromosome inactivation triggers fusion of A/B compartments to inactive X (Xi)-specific structures known as S1 and S2 compartments. SMCHD1 then merges S1/S2s to form the Xi super-structure. Here, we ask how S1/S2 compartments form and reveal that Xist RNA drives their formation via recruitment of Polycomb repressive complex 1 (PRC1). Ablating Smchd1 in post-XCI cells unveils S1/S2 structures. Loss of SMCHD1 leads to trapping Xist in the S1 compartment, impairing RNA spreading into S2. On the other hand, depleting Xist, PRC1, or HNRNPK precludes re-emergence of S1/S2 structures, and loss of S1/S2 compartments paradoxically strengthens the partition between Xi megadomains. Finally, Xi-reactivation in post-XCI cells can be enhanced by depleting both SMCHD1 and DNA methylation. We conclude that Xist, PRC1, and SMCHD1 collaborate in an obligatory, sequential manner to partition, fuse, and direct self-association of Xi compartments required for proper spreading of Xist RNA.
Conflict of interest statement
J.T.L. is a co-founder of and a scientific advisor for Translate Bio and Fulcrum Therapeutics. The other authors declare no competing interests.
Figures



Similar articles
-
SMCHD1 Merges Chromosome Compartments and Assists Formation of Super-Structures on the Inactive X.Cell. 2018 Jul 12;174(2):406-421.e25. doi: 10.1016/j.cell.2018.05.007. Epub 2018 Jun 7. Cell. 2018. PMID: 29887375 Free PMC article.
-
Smchd1 Targeting to the Inactive X Is Dependent on the Xist-HnrnpK-PRC1 Pathway.Cell Rep. 2018 Nov 13;25(7):1912-1923.e9. doi: 10.1016/j.celrep.2018.10.044. Cell Rep. 2018. PMID: 30428357
-
Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X.Mol Cell. 2019 Apr 4;74(1):101-117.e10. doi: 10.1016/j.molcel.2019.01.015. Epub 2019 Feb 28. Mol Cell. 2019. PMID: 30827740 Free PMC article.
-
Polycomb complexes in X chromosome inactivation.Philos Trans R Soc Lond B Biol Sci. 2017 Nov 5;372(1733):20170021. doi: 10.1098/rstb.2017.0021. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28947664 Free PMC article. Review.
-
A lifelong duty: how Xist maintains the inactive X chromosome.Curr Opin Genet Dev. 2022 Aug;75:101927. doi: 10.1016/j.gde.2022.101927. Epub 2022 Jun 16. Curr Opin Genet Dev. 2022. PMID: 35717799 Free PMC article. Review.
Cited by
-
Application of Hi-C and other omics data analysis in human cancer and cell differentiation research.Comput Struct Biotechnol J. 2021 Apr 8;19:2070-2083. doi: 10.1016/j.csbj.2021.04.016. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33995903 Free PMC article. Review.
-
Xist nucleates local protein gradients to propagate silencing across the X chromosome.Cell. 2021 Dec 9;184(25):6174-6192.e32. doi: 10.1016/j.cell.2021.10.022. Epub 2021 Nov 4. Cell. 2021. PMID: 34813726 Free PMC article.
-
Forged by DXZ4, FIRRE, and ICCE: How Tandem Repeats Shape the Active and Inactive X Chromosome.Front Cell Dev Biol. 2020 Jan 21;7:328. doi: 10.3389/fcell.2019.00328. eCollection 2019. Front Cell Dev Biol. 2020. PMID: 32076600 Free PMC article.
-
Long Non-Coding RNA (lncRNA) Roles in Cell Biology, Neurodevelopment and Neurological Disorders.Noncoding RNA. 2021 Jun 17;7(2):36. doi: 10.3390/ncrna7020036. Noncoding RNA. 2021. PMID: 34204536 Free PMC article. Review.
-
Traditional Banxia Xiexin decoction inhibits invasion, metastasis, and epithelial mesenchymal transition in gastric cancer by reducing lncRNA TUC338 expression.Heliyon. 2023 Oct 20;9(11):e21064. doi: 10.1016/j.heliyon.2023.e21064. eCollection 2023 Nov. Heliyon. 2023. PMID: 37964840 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

