A conserved RNA structure is essential for a satellite RNA-mediated inhibition of helper virus accumulation
- PMID: 31269212
- PMCID: PMC6735963
- DOI: 10.1093/nar/gkz564
A conserved RNA structure is essential for a satellite RNA-mediated inhibition of helper virus accumulation
Abstract
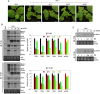
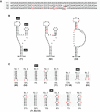
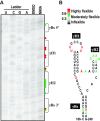
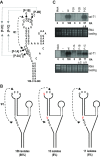
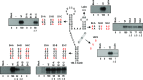
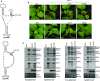
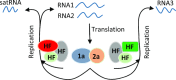
As a class of parasitic, non-coding RNAs, satellite RNAs (satRNAs) have to compete with their helper virus for limited amounts of viral and/or host resources for efficient replication, by which they usually reduce viral accumulation and symptom expression. Here, we report a cucumber mosaic virus (CMV)-associated satRNA (sat-T1) that ameliorated CMV-induced symptoms, accompanied with a significant reduction in the accumulation of viral genomic RNAs 1 and 2, which encode components of the viral replicase. Intrans replication assays suggest that the reduced accumulation is the outcome of replication competition. The structural basis of sat-T1 responsible for the inhibition of viral RNA accumulation was determined to be a three-way branched secondary structure that contains two biologically important hairpins. One is indispensable for the helper virus inhibition, and the other engages in formation of a tertiary pseudoknot structure that is essential for sat-T1 survival. The secondary structure containing the pseudoknot is the first RNA element with a biological phenotype experimentally identified in CMV satRNAs, and it is structurally conserved in most CMV satRNAs. Thus, this may be a generic method for CMV satRNAs to inhibit the accumulation of the helper virus via the newly-identified RNA structure.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Similar articles
-
Cucumber mosaic virus satellite RNAs that induce similar symptoms in melon plants show large differences in fitness.J Gen Virol. 2011 Aug;92(Pt 8):1930-1938. doi: 10.1099/vir.0.032359-0. Epub 2011 May 11. J Gen Virol. 2011. PMID: 21562122
-
Repair of the 3' proximal and internal deletions of a satellite RNA associated with Cucumber mosaic virus is directed toward restoring structural integrity.Virology. 2014 Feb;450-451:222-32. doi: 10.1016/j.virol.2013.12.008. Epub 2014 Jan 7. Virology. 2014. PMID: 24503085
-
Mapping helper virus functions for cucumber mosaic virus satellite RNA with pseudorecombinants derived from cucumber mosaic and tomato aspermy viruses.Virology. 1994 Dec;205(2):574-7. doi: 10.1006/viro.1994.1682. Virology. 1994. PMID: 7526544
-
Cucumoviruses.Adv Virus Res. 2003;62:241-323. doi: 10.1016/s0065-3527(03)62005-1. Adv Virus Res. 2003. PMID: 14719367 Review.
-
Satellite RNA pathogens of plants: impacts and origins-an RNA silencing perspective.Wiley Interdiscip Rev RNA. 2016 Jan-Feb;7(1):5-16. doi: 10.1002/wrna.1311. Epub 2015 Oct 20. Wiley Interdiscip Rev RNA. 2016. PMID: 26481458 Review.
Cited by
-
High-Throughput Sequencing Discloses the Cucumber Mosaic Virus (CMV) Diversity in Slovakia and Reveals New Hosts of CMV from the Papaveraceae Family.Plants (Basel). 2022 Jun 23;11(13):1665. doi: 10.3390/plants11131665. Plants (Basel). 2022. PMID: 35807616 Free PMC article.
-
Legacy of Plant Virology in Croatia-From Virus Identification to Molecular Epidemiology, Evolution, Genomics and Beyond.Viruses. 2021 Nov 23;13(12):2339. doi: 10.3390/v13122339. Viruses. 2021. PMID: 34960609 Free PMC article. Review.
-
The non-template functions of helper virus RNAs create optimal replication conditions to enhance the proliferation of satellite RNAs.PLoS Pathog. 2024 Apr 17;20(4):e1012174. doi: 10.1371/journal.ppat.1012174. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38630801 Free PMC article.
-
A special satellite-like RNA of a novel hypovirus from Pestalotiopsis fici broadens the definition of fungal satellite.PLoS Pathog. 2023 Jun 7;19(6):e1010889. doi: 10.1371/journal.ppat.1010889. eCollection 2023 Jun. PLoS Pathog. 2023. PMID: 37285391 Free PMC article.
-
Viroids, Satellite RNAs and Prions: Folding of Nucleic Acids and Misfolding of Proteins.Viruses. 2024 Feb 26;16(3):360. doi: 10.3390/v16030360. Viruses. 2024. PMID: 38543726 Free PMC article.
References
-
- Quinn J.J., Chang H.Y.. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016; 17:47–62. - PubMed
-
- Mercer T.R., Dinger M.E., Mattick J.S.. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009; 10:155–159. - PubMed
-
- Ponting C.P., Oliver P.L., Reik W.. Evolution and functions of long noncoding RNAs. Cell. 2009; 136:629–641. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

