GAIL: An interactive webserver for inference and dynamic visualization of gene-gene associations based on gene ontology guided mining of biomedical literature
- PMID: 31260503
- PMCID: PMC6602258
- DOI: 10.1371/journal.pone.0219195
GAIL: An interactive webserver for inference and dynamic visualization of gene-gene associations based on gene ontology guided mining of biomedical literature
Abstract
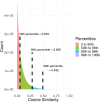
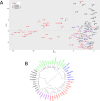
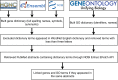
In systems biology, inference of functional associations among genes is compelling because the construction of functional association networks facilitates biomarker discovery. Specifically, such gene associations in human can help identify putative biomarkers that can be used as diagnostic tools in treating patients. Although biomedical literature is considered a valuable data source for this task, currently only a limited number of webservers are available for mining gene-gene associations from the vast amount of biomedical literature using text mining techniques. Moreover, these webservers often have limited coverage of biomedical literature and also lack efficient and user-friendly tools to interpret and visualize mined relationships among genes. To address these limitations, we developed GAIL (Gene-gene Association Inference based on biomedical Literature), an interactive webserver that infers human gene-gene associations from Gene Ontology (GO) guided biomedical literature mining and provides dynamic visualization of the resulting association networks and various gene set enrichment analysis tools. We evaluate the utility and performance of GAIL with applications to gene signatures associated with systemic lupus erythematosus and breast cancer. Results show that GAIL allows effective interrogation and visualization of gene-gene networks and their subnetworks, which facilitates biological understanding of gene-gene associations. GAIL is available at http://chunglab.io/GAIL/.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
PALMER: improving pathway annotation based on the biomedical literature mining with a constrained latent block model.BMC Bioinformatics. 2020 Oct 2;21(1):432. doi: 10.1186/s12859-020-03756-3. BMC Bioinformatics. 2020. PMID: 33008309 Free PMC article.
-
A statistical framework for biomedical literature mining.Stat Med. 2017 Sep 30;36(22):3461-3474. doi: 10.1002/sim.7384. Epub 2017 Jul 4. Stat Med. 2017. PMID: 28675924 Free PMC article.
-
LiverCancerMarkerRIF: a liver cancer biomarker interactive curation system combining text mining and expert annotations.Database (Oxford). 2014 Aug 27;2014:bau085. doi: 10.1093/database/bau085. Print 2014. Database (Oxford). 2014. PMID: 25168057 Free PMC article.
-
Analysis of biological processes and diseases using text mining approaches.Methods Mol Biol. 2010;593:341-82. doi: 10.1007/978-1-60327-194-3_16. Methods Mol Biol. 2010. PMID: 19957157 Review.
-
How to learn about gene function: text-mining or ontologies?Methods. 2015 Mar;74:3-15. doi: 10.1016/j.ymeth.2014.07.004. Epub 2014 Aug 1. Methods. 2015. PMID: 25088781 Review.
Cited by
-
PALMER: improving pathway annotation based on the biomedical literature mining with a constrained latent block model.BMC Bioinformatics. 2020 Oct 2;21(1):432. doi: 10.1186/s12859-020-03756-3. BMC Bioinformatics. 2020. PMID: 33008309 Free PMC article.
-
Identification and validation of a prognostic signature and combination drug therapy for immunotherapy of head and neck squamous cell carcinoma.Comput Struct Biotechnol J. 2021 Feb 9;19:1263-1276. doi: 10.1016/j.csbj.2021.01.046. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33717423 Free PMC article.
-
Sparse Linear Discriminant Analysis using the Prior-Knowledge-Guided Block Covariance Matrix.Chemometr Intell Lab Syst. 2020 Nov 15;206:104142. doi: 10.1016/j.chemolab.2020.104142. Epub 2020 Aug 27. Chemometr Intell Lab Syst. 2020. PMID: 32968333 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

