Transcriptomic Analysis of Aedes aegypti Innate Immune System in Response to Ingestion of Chikungunya Virus
- PMID: 31252518
- PMCID: PMC6651163
- DOI: 10.3390/ijms20133133
Transcriptomic Analysis of Aedes aegypti Innate Immune System in Response to Ingestion of Chikungunya Virus
Abstract
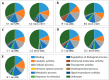
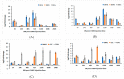
Aedes aegypti (L.) is the primary vector of emergent mosquito-borne viruses, including chikungunya, dengue, yellow fever, and Zika viruses. To understand how these viruses interact with their mosquito vectors, an analysis of the innate immune system response was conducted. The innate immune system is a conserved evolutionary defense strategy and is the dominant immune system response found in invertebrates and vertebrates, as well as plants. RNA-sequencing analysis was performed to compare target transcriptomes of two Florida Ae. aegypti strains in response to chikungunya virus infection. We analyzed a strain collected from a field population in Key West, Florida, and a laboratory strain originating from Orlando. A total of 1835 transcripts were significantly expressed at different levels between the two Florida strains of Ae. aegypti. Gene Ontology analysis placed these genes into 12 categories of biological processes, including 856 transcripts (up/down regulated) with more than 1.8-fold (p-adj (p-adjust value) ≤ 0.01). Transcriptomic analysis and q-PCR data indicated that the members of the AaeCECH genes are important for chikungunya infection response in Ae. aegypti. These immune-related enzymes that the chikungunya virus infection induces may inform molecular-based strategies for interruption of arbovirus transmission by mosquitoes.
Keywords: Aedes aegypti; anti-microbial peptide; chikungunya virus; gene expression; immune responses; transcriptome.
Conflict of interest statement
All authors declare no conflict of interest.
Figures
Similar articles
-
Transcription Profiling for Defensins of Aedes aegypti (Diptera: Culicidae) During Development and in Response to Infection With Chikungunya and Zika Viruses.J Med Entomol. 2018 Jan 10;55(1):78-89. doi: 10.1093/jme/tjx174. J Med Entomol. 2018. PMID: 28968775
-
Transcriptional Profile of Aedes aegypti Leucine-Rich Repeat Proteins in Response to Zika and Chikungunya Viruses.Int J Mol Sci. 2019 Jan 31;20(3):615. doi: 10.3390/ijms20030615. Int J Mol Sci. 2019. PMID: 30708982 Free PMC article.
-
The Effect of Permethrin Resistance on Aedes aegypti Transcriptome Following Ingestion of Zika Virus Infected Blood.Viruses. 2018 Sep 1;10(9):470. doi: 10.3390/v10090470. Viruses. 2018. PMID: 30200481 Free PMC article.
-
The immune strategies of mosquito Aedes aegypti against microbial infection.Dev Comp Immunol. 2018 Jun;83:12-21. doi: 10.1016/j.dci.2017.12.001. Epub 2017 Dec 5. Dev Comp Immunol. 2018. PMID: 29217264 Review.
-
Endogenous non-retroviral elements in genomes of Aedes mosquitoes and vector competence.Emerg Microbes Infect. 2019;8(1):542-555. doi: 10.1080/22221751.2019.1599302. Emerg Microbes Infect. 2019. PMID: 30938223 Free PMC article. Review.
Cited by
-
Ross River Virus Provokes Differentially Expressed MicroRNA and RNA Interference Responses in Aedes aegypti Mosquitoes.Viruses. 2020 Jun 27;12(7):695. doi: 10.3390/v12070695. Viruses. 2020. PMID: 32605094 Free PMC article.
-
A Review of Omics Studies on Arboviruses: Alphavirus, Orthobunyavirus and Phlebovirus.Viruses. 2022 Oct 5;14(10):2194. doi: 10.3390/v14102194. Viruses. 2022. PMID: 36298749 Free PMC article. Review.
-
A comprehensive review of Wolbachia-mediated mechanisms to control dengue virus transmission in Aedes aegypti through innate immune pathways.Front Immunol. 2024 Aug 8;15:1434003. doi: 10.3389/fimmu.2024.1434003. eCollection 2024. Front Immunol. 2024. PMID: 39176079 Free PMC article. Review.
-
Defining the mechanisms of action and mosquito larva midgut response to a yeast-encapsulated orange oil larvicide.Parasit Vectors. 2022 May 28;15(1):183. doi: 10.1186/s13071-022-05307-6. Parasit Vectors. 2022. PMID: 35643588 Free PMC article.
-
Transcriptome Analysis of Response to Zika Virus Infection in Two Aedes albopictus Strains with Different Vector Competence.Int J Mol Sci. 2023 Feb 21;24(5):4257. doi: 10.3390/ijms24054257. Int J Mol Sci. 2023. PMID: 36901688 Free PMC article.
References
-
- Simo Tchetgna H., Sem Ouilibona R., Nkili-Meyong A.A., Caron M., Labouba I., Selekon B., Njouom R., Leroy E.M., Nakoune E., Berthet N. Viral Exploration of Negative Acute Febrile Cases Observed during Chikungunya Outbreaks in Gabon. Intervirology. 2018;61:174–184. doi: 10.1159/000495136. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

