A Tour de Force on the Double Helix: Exploiting DNA Mechanics To Study DNA-Based Molecular Machines
- PMID: 31251042
- PMCID: PMC6879785
- DOI: 10.1021/acs.biochem.9b00346
A Tour de Force on the Double Helix: Exploiting DNA Mechanics To Study DNA-Based Molecular Machines
Abstract
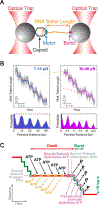
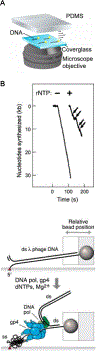
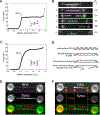
DNA is both a fundamental building block of life and a fascinating natural polymer. The advent of single-molecule manipulation tools made it possible to exert controlled force on individual DNA molecules and measure their mechanical response. Such investigations elucidated the elastic properties of DNA and revealed its distinctive structural configurations across force regimes. In the meantime, a detailed understanding of DNA mechanics laid the groundwork for single-molecule studies of DNA-binding proteins and DNA-processing enzymes that bend, stretch, and twist DNA. These studies shed new light on the metabolism and transactions of nucleic acids, which constitute a major part of the cell's operating system. Furthermore, the marriage of single-molecule fluorescence visualization and force manipulation has enabled researchers to directly correlate the applied tension to changes in the DNA structure and the behavior of DNA-templated complexes. Overall, experimental exploitation of DNA mechanics has been and will continue to be a unique and powerful strategy for understanding how molecular machineries recognize and modify the physical state of DNA to accomplish their biological functions.
Figures
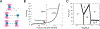
Similar articles
-
Recent developments in single-molecule DNA mechanics.Curr Opin Struct Biol. 2012 Jun;22(3):304-12. doi: 10.1016/j.sbi.2012.04.007. Epub 2012 May 31. Curr Opin Struct Biol. 2012. PMID: 22658779 Free PMC article. Review.
-
Hyperstretching DNA.Nat Commun. 2017 Dec 19;8(1):2197. doi: 10.1038/s41467-017-02396-1. Nat Commun. 2017. PMID: 29259297 Free PMC article.
-
Molecular scaffolds: when DNA becomes the hardware for single-molecule investigations.Curr Opin Chem Biol. 2019 Dec;53:192-203. doi: 10.1016/j.cbpa.2019.09.006. Epub 2019 Nov 20. Curr Opin Chem Biol. 2019. PMID: 31759266 Review.
-
When Force Met Fluorescence: Single-Molecule Manipulation and Visualization of Protein-DNA Interactions.Annu Rev Biophys. 2024 Jul;53(1):169-191. doi: 10.1146/annurev-biophys-030822-032904. Epub 2024 Jun 28. Annu Rev Biophys. 2024. PMID: 38237015 Review.
-
Integrated magnetic tweezers and single-molecule FRET for investigating the mechanical properties of nucleic acid.Methods. 2016 Aug 1;105:16-25. doi: 10.1016/j.ymeth.2016.06.009. Epub 2016 Jun 15. Methods. 2016. PMID: 27320203 Free PMC article.
Cited by
-
Smc5/6's multifaceted DNA binding capacities stabilize branched DNA structures.Nat Commun. 2022 Nov 23;13(1):7179. doi: 10.1038/s41467-022-34928-9. Nat Commun. 2022. PMID: 36418314 Free PMC article.
-
Probing the Interaction Between Chromatin and Chromatin-Associated Complexes with Optical Tweezers.Methods Mol Biol. 2022;2478:313-327. doi: 10.1007/978-1-0716-2229-2_11. Methods Mol Biol. 2022. PMID: 36063325 Free PMC article.
-
Looking at Biomolecular Interactions through the Lens of Correlated Fluorescence Microscopy and Optical Tweezers.Int J Mol Sci. 2023 Jan 31;24(3):2668. doi: 10.3390/ijms24032668. Int J Mol Sci. 2023. PMID: 36768987 Free PMC article. Review.
-
Modeling multiple duplex DNA attachments in a force-extension experiment.Biophys Rep (N Y). 2022 Feb 2;2(1):100045. doi: 10.1016/j.bpr.2022.100045. eCollection 2022 Mar 9. Biophys Rep (N Y). 2022. PMID: 36425083 Free PMC article.
References
-
- Watson JD, and Crick FH (1953) Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid, Nature 171, 737–738. - PubMed
-
- Wilkins MH, Stokes AR, and Wilson HR (1953) Molecular structure of deoxypentose nucleic acids, Nature 171, 738–740. - PubMed
-
- Franklin RE, and Gosling RG (1953) Molecular configuration in sodium thymonucleate, Nature 171, 740–741. - PubMed
-
- Griffith J, Huberman JA, and Kornberg A (1971) Electron microscopy of DNA polymerase bound to DNA, Journal of moìecuìar biology 55, 209–214. - PubMed
-
- Morikawa K, and Yanagida M (1981) Visualization of individual DNA molecules in solution by light microscopy: DAPI staining method, Journal of biochemistry 89, 693–696. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

