Fndc-1 contributes to paternal mitochondria elimination in C. elegans
- PMID: 31233739
- PMCID: PMC6717525
- DOI: 10.1016/j.ydbio.2019.06.016
Fndc-1 contributes to paternal mitochondria elimination in C. elegans
Abstract
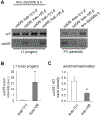
Paternal mitochondria are eliminated following fertilization by selective autophagy, but the mechanisms that restrict this process to sperm-derived organelles are not well understood. FUNDC1 (FUN14 domain containing 1) is a mammalian mitophagy receptor expressed on the mitochondrial outer membrane that contributes to mitochondrial quality control following hypoxic stress. Like FUNDC1, the C. elegans ortholog FNDC-1 is widely expressed in somatic tissues and mediates hypoxic mitophagy. Here, we report that FNDC-1 is strongly expressed in sperm but not oocytes and contributes to paternal mitochondria elimination. Paternal mitochondrial DNA is normally undetectable in wildtype larva, but can be detected in the cross-progeny of fndc-1 mutant males. Moreover, loss of fndc-1 retards the rate of paternal mitochondria degradation, but not that of membranous organelles, a nematode specific membrane compartment whose fusion is required for sperm motility. This is the first example of a ubiquitin-independent mitophagy receptor playing a role in the selective degradation of sperm mitochondria.
Keywords: Autophagy; C. elegans; Maternal inheritance; Mitochondria; Mitophagy.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures
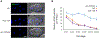

Similar articles
-
Ubiquitination is required for the initial removal of paternal organelles in C. elegans.Dev Biol. 2019 Sep 15;453(2):168-179. doi: 10.1016/j.ydbio.2019.05.015. Epub 2019 May 30. Dev Biol. 2019. PMID: 31153831 Free PMC article.
-
Autophagosomal Sperm Organelle Clearance and mtDNA Inheritance in C. elegans.Adv Anat Embryol Cell Biol. 2019;231:1-23. doi: 10.1007/102_2018_1. Adv Anat Embryol Cell Biol. 2019. PMID: 30467692 Review.
-
Dual roles for ubiquitination in the processing of sperm organelles after fertilization.BMC Dev Biol. 2014 Feb 15;14:6. doi: 10.1186/1471-213X-14-6. BMC Dev Biol. 2014. PMID: 24528894 Free PMC article.
-
Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos.Science. 2011 Nov 25;334(6059):1141-4. doi: 10.1126/science.1210333. Epub 2011 Oct 13. Science. 2011. PMID: 21998252
-
Mitophagy during development and stress in C. elegans.Mech Ageing Dev. 2020 Jul;189:111266. doi: 10.1016/j.mad.2020.111266. Epub 2020 May 23. Mech Ageing Dev. 2020. PMID: 32454052 Review.
Cited by
-
Effects of reactive oxygen species and mitochondrial dysfunction on reproductive aging.Front Cell Dev Biol. 2024 Feb 23;12:1347286. doi: 10.3389/fcell.2024.1347286. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38465288 Free PMC article. Review.
-
Deletion of FUNDC2 and CMC4 on Chromosome Xq28 Is Sufficient to Cause Hypergonadotropic Hypogonadism in Men.Front Genet. 2020 Sep 22;11:557341. doi: 10.3389/fgene.2020.557341. eCollection 2020. Front Genet. 2020. PMID: 33193636 Free PMC article.
-
Molecular Mechanisms and Regulation of Mammalian Mitophagy.Cells. 2021 Dec 23;11(1):38. doi: 10.3390/cells11010038. Cells. 2021. PMID: 35011599 Free PMC article. Review.
-
Mitophagy of polarized sperm-derived mitochondria after fertilization.iScience. 2020 Dec 31;24(1):102029. doi: 10.1016/j.isci.2020.102029. eCollection 2021 Jan 22. iScience. 2020. PMID: 33506190 Free PMC article.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1.Autophagy. 2021 Jan;17(1):1-382. doi: 10.1080/15548627.2020.1797280. Epub 2021 Feb 8. Autophagy. 2021. PMID: 33634751 Free PMC article.
References
-
- Sato M, Sato K (2011) Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334: 1141–1144. - PubMed
-
- Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V (2011) Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334: 1144–1147. - PubMed
-
- Whitworth AJ, Pallanck LJ (2017) PINK1/Parkin mitophagy and neurodegeneration-what do we really know in vivo? Curr Opin Genet Dev 44: 47–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

