Identification of Immunodominant HIV-1 Epitopes Presented by HLA-C*12:02, a Protective Allele, Using an Immunopeptidomics Approach
- PMID: 31217245
- PMCID: PMC6694829
- DOI: 10.1128/JVI.00634-19
Identification of Immunodominant HIV-1 Epitopes Presented by HLA-C*12:02, a Protective Allele, Using an Immunopeptidomics Approach
Abstract
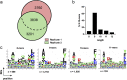
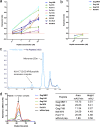
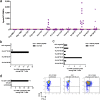
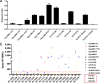
Despite the fact that the cell surface expression level of HLA-C on both uninfected and HIV-infected cells is lower than those of HLA-A and -B, increasing evidence suggests an important role for HLA-C and HLA-C-restricted CD8+ T cell responses in determining the efficiency of viral control in HIV-1-infected individuals. Nonetheless, HLA-C-restricted T cell responses are much less well studied than HLA-A/B-restricted ones, and relatively few optimal HIV-1 CD8+ T cell epitopes restricted by HLA-C alleles have been defined. Recent improvements in the sensitivity of mass spectrometry (MS)-based approaches for profiling the immunopeptidome present an opportunity for epitope discovery on a large scale. Here, we employed an MS-based immunopeptidomic strategy to characterize HIV-1 peptides presented by a protective allele, HLA-C*12:02. We identified a total of 10,799 unique 8- to 12-mer peptides, including 15 HIV-1 peptides. The latter included 2 previously reported immunodominant HIV-1 epitopes, and analysis of T cell responses to the other HIV-1 peptides detected revealed an additional immunodominant epitope. These findings illustrate the utility of MS-based approaches for epitope definition and emphasize the capacity of HLA-C to present immunodominant T cell epitopes in HIV-infected individuals, indicating the importance of further evaluation of HLA-C-restricted responses to identify novel targets for HIV-1 prophylactic and therapeutic strategies.IMPORTANCE Mass spectrometry (MS)-based approaches are increasingly being employed for large-scale identification of HLA-bound peptides derived from pathogens, but only very limited profiling of the HIV-1 immunopeptidome has been conducted to date. Notably, a growing body of evidence has recently begun to indicate a protective role for HLA-C in HIV-1 infection, which may suggest that despite the fact that levels of HLA-C expression on both uninfected and HIV-1-infected cells are lower than those of HLA-A/B, HLA-C still presents epitopes to CD8+ T cells effectively. To explore this, we analyzed HLA-C*12:02-restricted HIV-1 peptides presented on HIV-1-infected cells expressing only HLA-C*12:02 (a protective allele) using liquid chromatography-tandem MS (LC-MS/MS). We identified a number of novel HLA-C*12:02-bound HIV-1 peptides and showed that although the majority of them did not elicit T cell responses during natural infection in a Japanese cohort, they included three immunodominant epitopes, emphasizing the contribution of HLA-C to epitope presentation on HIV-infected cells.
Keywords: CTL; HIV-1; HLA-C; LC-MS/MS; epitope; mass spectrometry; peptide.
Copyright © 2019 Chikata et al.
Figures


Similar articles
-
Impact of Micropolymorphism Outside the Peptide Binding Groove in the Clinically Relevant Allele HLA-C*14 on T Cell Responses in HIV-1 Infection.J Virol. 2022 May 25;96(10):e0043222. doi: 10.1128/jvi.00432-22. Epub 2022 Apr 27. J Virol. 2022. PMID: 35475667 Free PMC article.
-
Discrimination Between Human Leukocyte Antigen Class I-Bound and Co-Purified HIV-Derived Peptides in Immunopeptidomics Workflows.Front Immunol. 2018 Apr 27;9:912. doi: 10.3389/fimmu.2018.00912. eCollection 2018. Front Immunol. 2018. PMID: 29780384 Free PMC article.
-
HIV-1-specific cytotoxic T lymphocyte (CTL) responses against immunodominant optimal epitopes slow the progression of AIDS in China.Curr HIV Res. 2008 Jun;6(4):335-50. doi: 10.2174/157016208785132473. Curr HIV Res. 2008. PMID: 18691032
-
Identification and selection of immunodominant B and T cell epitopes for dengue multi-epitope-based vaccine.Med Microbiol Immunol. 2021 Feb;210(1):1-11. doi: 10.1007/s00430-021-00700-x. Epub 2021 Jan 30. Med Microbiol Immunol. 2021. PMID: 33515283 Review.
-
T Cell Epitope Discovery in the Context of Distinct and Unique Indigenous HLA Profiles.Front Immunol. 2022 May 6;13:812393. doi: 10.3389/fimmu.2022.812393. eCollection 2022. Front Immunol. 2022. PMID: 35603215 Free PMC article. Review.
Cited by
-
IFNγ Modulates the Immunopeptidome of Triple Negative Breast Cancer Cells by Enhancing and Diversifying Antigen Processing and Presentation.Front Immunol. 2021 Apr 22;12:645770. doi: 10.3389/fimmu.2021.645770. eCollection 2021. Front Immunol. 2021. PMID: 33968037 Free PMC article.
-
STING Ligand-Mediated Priming of Functional CD8+ T Cells Specific for HIV-1-Protective Epitopes from Naive T Cells.J Virol. 2021 Jul 26;95(16):e0069921. doi: 10.1128/JVI.00699-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34076478 Free PMC article.
-
A Systematic Review of T Cell Epitopes Defined from the Proteome of Hepatitis B Virus.Vaccines (Basel). 2022 Feb 8;10(2):257. doi: 10.3390/vaccines10020257. Vaccines (Basel). 2022. PMID: 35214714 Free PMC article. Review.
-
Epitope-dependent effect of long-term cART on maintenance and recovery of HIV-1-specific CD8+ T cells.J Virol. 2023 Nov 30;97(11):e0102423. doi: 10.1128/jvi.01024-23. Epub 2023 Oct 25. J Virol. 2023. PMID: 37877716 Free PMC article.
-
Leveraging Immunopeptidomics To Study and Combat Infectious Disease.mSystems. 2021 Aug 31;6(4):e0031021. doi: 10.1128/mSystems.00310-21. Epub 2021 Aug 3. mSystems. 2021. PMID: 34342538 Free PMC article.
References
-
- Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527. doi:10.1038/nature10003. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

