A Unified Model for Inclusive Inheritance in Livestock Species
- PMID: 31209104
- PMCID: PMC6707455
- DOI: 10.1534/genetics.119.302375
A Unified Model for Inclusive Inheritance in Livestock Species
Abstract
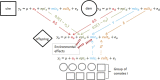
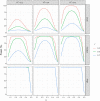
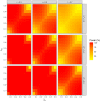
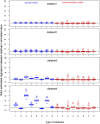
For years, animal selection in livestock species has been performed by selecting animals based on genetic inheritance. However, evolutionary studies have reported that nongenetic information that drives natural selection can also be inherited across generations (epigenetic, microbiota, environmental inheritance). In response to this finding, the concept of inclusive heritability, which combines all sources of information inherited across generations, was developed. To better predict the transmissible potential of each animal by taking into account these diverse sources of inheritance and improve selection in livestock species, we propose the "transmissibility model." Similarly to the animal model, this model uses pedigree and phenotypic information to estimate variance components and predict the transmissible potential of an individual, but differs by estimating the path coefficients of inherited information from parent to offspring instead of using a set value of 0.5 for both the sire and the dam (additive genetic relationship matrix). We demonstrated the structural identifiability of the transmissibility model, and performed a practical identifiability and power study of the model. We also performed simulations to compare the performances of the animal and transmissibility models for estimating the covariances between relatives and predicting the transmissible potential under different combinations of sources of inheritance. The transmissibility model provided similar results to the animal model when inheritance was of genetic origin only, but outperformed the animal model for estimating the covariances between relatives and predicting the transmissible potential when the proportion of inheritance of nongenetic origin was high or when the sire and dam path coefficients were very different.
Keywords: inclusive inheritance; livestock; model.
Copyright © 2019 by the Genetics Society of America.
Figures


Similar articles
-
An improved transmissibility model to detect transgenerational transmitted environmental effects.Genet Sel Evol. 2023 Sep 21;55(1):66. doi: 10.1186/s12711-023-00833-y. Genet Sel Evol. 2023. PMID: 37735633 Free PMC article.
-
Inclusive inheritance for residual feed intake in pigs and rabbits.J Anim Breed Genet. 2020 Nov;137(6):535-544. doi: 10.1111/jbg.12494. Epub 2020 Jul 22. J Anim Breed Genet. 2020. PMID: 32697021 Free PMC article.
-
Intergenerational Transmission of Characters Through Genetics, Epigenetics, Microbiota, and Learning in Livestock.Front Genet. 2019 Oct 31;10:1058. doi: 10.3389/fgene.2019.01058. eCollection 2019. Front Genet. 2019. PMID: 31737041 Free PMC article. Review.
-
Potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs.Genet Sel Evol. 2015 Jul 2;47(1):55. doi: 10.1186/s12711-015-0135-3. Genet Sel Evol. 2015. PMID: 26133579 Free PMC article.
-
Complex Trait Prediction from Genome Data: Contrasting EBV in Livestock to PRS in Humans: Genomic Prediction.Genetics. 2019 Apr;211(4):1131-1141. doi: 10.1534/genetics.119.301859. Genetics. 2019. PMID: 30967442 Free PMC article. Review.
Cited by
-
The potential of microbiota information to better predict efficiency traits in growing pigs fed a conventional and a high-fiber diet.Genet Sel Evol. 2024 Jan 19;56(1):8. doi: 10.1186/s12711-023-00865-4. Genet Sel Evol. 2024. PMID: 38243193 Free PMC article.
-
An improved transmissibility model to detect transgenerational transmitted environmental effects.Genet Sel Evol. 2023 Sep 21;55(1):66. doi: 10.1186/s12711-023-00833-y. Genet Sel Evol. 2023. PMID: 37735633 Free PMC article.
-
Gut microbiota and host genetics contribute to the phenotypic variation of digestive and feed efficiency traits in growing pigs fed a conventional and a high fiber diet.Genet Sel Evol. 2022 Jul 27;54(1):55. doi: 10.1186/s12711-022-00742-6. Genet Sel Evol. 2022. PMID: 35896976 Free PMC article.
-
Mapping the past, present and future research landscape of paternal effects.BMC Biol. 2020 Nov 27;18(1):183. doi: 10.1186/s12915-020-00892-3. BMC Biol. 2020. PMID: 33246472 Free PMC article.
-
Inclusive inheritance for residual feed intake in pigs and rabbits.J Anim Breed Genet. 2020 Nov;137(6):535-544. doi: 10.1111/jbg.12494. Epub 2020 Jul 22. J Anim Breed Genet. 2020. PMID: 32697021 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources

