A Series of Fortunate Events: Introducing Chlamydomonas as a Reference Organism
- PMID: 31189738
- PMCID: PMC6713297
- DOI: 10.1105/tpc.18.00952
A Series of Fortunate Events: Introducing Chlamydomonas as a Reference Organism
Abstract
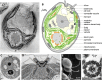
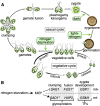
The unicellular alga Chlamydomonas reinhardtii is a classical reference organism for studying photosynthesis, chloroplast biology, cell cycle control, and cilia structure and function. It is also an emerging model for studying sensory cilia, the production of high-value bioproducts, and in situ structural determination. Much of the early appeal of Chlamydomonas was rooted in its promise as a genetic system, but like other classic model organisms, this rise to prominence predated the discovery of the structure of DNA, whole-genome sequences, and molecular techniques for gene manipulation. The haploid genome of C. reinhardtii facilitates genetic analyses and offers many of the advantages of microbial systems applied to a photosynthetic organism. C. reinhardtii has contributed to our understanding of chloroplast-based photosynthesis and cilia biology. Despite pervasive transgene silencing, technological advances have allowed researchers to address outstanding lines of inquiry in algal research. The most thoroughly studied unicellular alga, C. reinhardtii, is the current standard for algal research, and although genome editing is still far from efficient and routine, it nevertheless serves as a template for other algae. We present a historical retrospective of the rise of C. reinhardtii to illuminate its past and present. We also present resources for current and future scientists who may wish to expand their studies to the realm of microalgae.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures



Similar articles
-
Functional genomics of plant photosynthesis in the fast lane using Chlamydomonas reinhardtii.Trends Plant Sci. 2001 Aug;6(8):364-71. doi: 10.1016/s1360-1385(01)02018-0. Trends Plant Sci. 2001. PMID: 11495790 Review.
-
Chlamydomonas: Fast tracking from genomics.J Phycol. 2023 Aug;59(4):644-652. doi: 10.1111/jpy.13356. Epub 2023 Jul 7. J Phycol. 2023. PMID: 37417760
-
Chlamydomonas reinhardtii as the photosynthetic yeast.Annu Rev Genet. 1995;29:209-30. doi: 10.1146/annurev.ge.29.120195.001233. Annu Rev Genet. 1995. PMID: 8825474 Review.
-
Tools and techniques for chloroplast transformation of Chlamydomonas.Adv Exp Med Biol. 2007;616:34-45. doi: 10.1007/978-0-387-75532-8_4. Adv Exp Med Biol. 2007. PMID: 18161489 Review.
-
From molecular manipulation of domesticated Chlamydomonas reinhardtii to survival in nature.Elife. 2018 Nov 1;7:e39233. doi: 10.7554/eLife.39233. Elife. 2018. PMID: 30382941 Free PMC article. Review.
Cited by
-
Rapid and Efficient Colony-PCR for High Throughput Screening of Genetically Transformed Chlamydomonas reinhardtii.Life (Basel). 2020 Sep 10;10(9):186. doi: 10.3390/life10090186. Life (Basel). 2020. PMID: 32927613 Free PMC article.
-
Macular pigment-enriched oil production from genome-edited microalgae.Microb Cell Fact. 2022 Feb 19;21(1):27. doi: 10.1186/s12934-021-01736-7. Microb Cell Fact. 2022. PMID: 35183173 Free PMC article.
-
Nourseothricin N-acetyl transferase (NAT), a new selectable marker for nuclear gene expression in Chlamydomonas.Plant Methods. 2019 Nov 19;15:140. doi: 10.1186/s13007-019-0526-5. eCollection 2019. Plant Methods. 2019. PMID: 31827577 Free PMC article.
-
Microfluidic Platforms Designed for Morphological and Photosynthetic Investigations of Chlamydomonas reinhardtii on a Single-Cell Level.Cells. 2022 Jan 14;11(2):285. doi: 10.3390/cells11020285. Cells. 2022. PMID: 35053401 Free PMC article.
-
Optimizing Fe Nutrition for Algal Growth.Methods Mol Biol. 2023;2665:203-215. doi: 10.1007/978-1-0716-3183-6_16. Methods Mol Biol. 2023. PMID: 37166603
References
-
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. - PubMed
-
- Asamizu E., Nakamura Y., Miura K., Fukuzawa H., Fujiwara S., Hirono M., Iwamoto K., Matsuda Y., Minagawa J., Shimogawara K., Takahashi Y., Tabata S. (2004). Establishment of publicly available cDNA material and information resource of Chlamydomonas reinhardtii (Chlorophyta) to facilitate gene function analysis. Phycologia 43: 722–726.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

