CHML promotes liver cancer metastasis by facilitating Rab14 recycle
- PMID: 31175290
- PMCID: PMC6555802
- DOI: 10.1038/s41467-019-10364-0
CHML promotes liver cancer metastasis by facilitating Rab14 recycle
Abstract
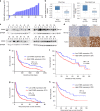
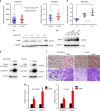
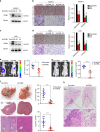
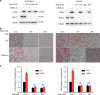
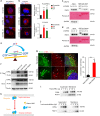
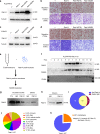
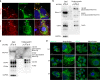
Metastasis-associated recurrence is the major cause of poor prognosis in hepatocellular carcinoma (HCC), however, the underlying mechanisms remain largely elusive. In this study, we report that expression of choroideremia-like (CHML) is increased in HCC, associated with poor survival, early recurrence and more satellite nodules in HCC patients. CHML promotes migration, invasion and metastasis of HCC cells, in a Rab14-dependent manner. Mechanism study reveals that CHML facilitates constant recycling of Rab14 by escorting Rab14 to the membrane. Furthermore, we identify several metastasis regulators as cargoes carried by Rab14-positive vesicles, including Mucin13 and CD44, which may contribute to metastasis-promoting effects of CHML. Altogether, our data establish CHML as a potential promoter of HCC metastasis, and the CHML-Rab14 axis may be a promising therapeutic target for HCC.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
MicroRNA-34c-3p promotes cell proliferation and invasion in hepatocellular carcinoma by regulation of NCKAP1 expression.J Cancer Res Clin Oncol. 2017 Feb;143(2):263-273. doi: 10.1007/s00432-016-2280-7. Epub 2016 Oct 4. J Cancer Res Clin Oncol. 2017. PMID: 27704267
-
Ack1 overexpression promotes metastasis and indicates poor prognosis of hepatocellular carcinoma.Oncotarget. 2015 Dec 1;6(38):40622-41. doi: 10.18632/oncotarget.5872. Oncotarget. 2015. PMID: 26536663 Free PMC article. Clinical Trial.
-
Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma.J Hepatol. 2012 Sep;57(3):600-12. doi: 10.1016/j.jhep.2012.04.020. Epub 2012 May 18. J Hepatol. 2012. PMID: 22613004
-
Overexpression of forkhead box C1 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma.Hepatology. 2013 Feb;57(2):610-24. doi: 10.1002/hep.26029. Hepatology. 2013. PMID: 22911555
-
Loss of androgen receptor promotes HCC invasion and metastasis via activating circ-LNPEP/miR-532-3p/RAB9A signal under hypoxia.Biochem Biophys Res Commun. 2021 Jun 11;557:26-32. doi: 10.1016/j.bbrc.2021.02.120. Epub 2021 Apr 13. Biochem Biophys Res Commun. 2021. PMID: 33862456
Cited by
-
The role of long non-coding RNA HCG18 in cancer.Clin Transl Oncol. 2023 Mar;25(3):611-619. doi: 10.1007/s12094-022-02992-8. Epub 2022 Nov 8. Clin Transl Oncol. 2023. PMID: 36346572 Review.
-
Prognostic signatures of sphingolipids: Understanding the immune landscape and predictive role in immunotherapy response and outcomes of hepatocellular carcinoma.Front Immunol. 2023 Mar 17;14:1153423. doi: 10.3389/fimmu.2023.1153423. eCollection 2023. Front Immunol. 2023. PMID: 37006285 Free PMC article.
-
The Dispensable Roles of X-Linked Ubl4a and Its Autosomal Counterpart Ubl4b in Spermatogenesis Represent a New Evolutionary Type of X-Derived Retrogenes.Front Genet. 2021 Jun 25;12:689902. doi: 10.3389/fgene.2021.689902. eCollection 2021. Front Genet. 2021. PMID: 34249105 Free PMC article.
-
Resveratrol inhibits the migration, invasion and epithelial-mesenchymal transition in liver cancer cells through up- miR-186-5p expression.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2021 Oct 25;50(5):582-590. doi: 10.3724/zdxbyxb-2021-0197. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2021. PMID: 34986537 Free PMC article. English.
-
The Effects of a Novel Curcumin Derivative Loaded Long-Circulating Solid Lipid Nanoparticle on the MHCC-97H Liver Cancer Cells and Pharmacokinetic Behavior.Int J Nanomedicine. 2022 May 17;17:2225-2241. doi: 10.2147/IJN.S363237. eCollection 2022. Int J Nanomedicine. 2022. PMID: 35607705 Free PMC article.
References
-
- Lochan, Reeves, H. L. & Manas, D. M. Surgical management of HCC. In: Practical Management of Chronic Viral Hepatitis (ed Serviddio, G.) (InTech, London, UK, 2013).
-
- Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers2, 16018 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

