Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction
- PMID: 31175275
- PMCID: PMC6555806
- DOI: 10.1038/s41467-019-10409-4
Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction
Abstract
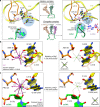
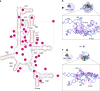
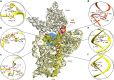
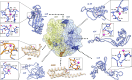
The ribosome, the largest RNA-containing macromolecular machinery in cells, requires metal ions not only to maintain its three-dimensional fold but also to perform protein synthesis. Despite the vast biochemical data regarding the importance of metal ions for efficient protein synthesis and the increasing number of ribosome structures solved by X-ray crystallography or cryo-electron microscopy, the assignment of metal ions within the ribosome remains elusive due to methodological limitations. Here we present extensive experimental data on the potassium composition and environment in two structures of functional ribosome complexes obtained by measurement of the potassium anomalous signal at the K-edge, derived from long-wavelength X-ray diffraction data. We elucidate the role of potassium ions in protein synthesis at the three-dimensional level, most notably, in the environment of the ribosome functional decoding and peptidyl transferase centers. Our data expand the fundamental knowledge of the mechanism of ribosome function and structural integrity.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Ribosome structure. The ribosome in action.Science. 2001 May 4;292(5518):868-9. doi: 10.1126/science.1061513. Science. 2001. PMID: 11341282 No abstract available.
-
Escherichia coli 70 S ribosome at 15 A resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein.J Mol Biol. 1998 Jul 3;280(1):103-16. doi: 10.1006/jmbi.1998.1859. J Mol Biol. 1998. PMID: 9653034
-
Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements.Cell. 2006 Sep 22;126(6):1065-77. doi: 10.1016/j.cell.2006.08.032. Epub 2006 Sep 7. Cell. 2006. PMID: 16962654
-
Deepening ribosomal insights.ACS Chem Biol. 2006 Oct 24;1(9):567-9. doi: 10.1021/cb600407u. ACS Chem Biol. 2006. PMID: 17168551 Review.
-
New Insights into Ribosome Structure and Function.Cold Spring Harb Perspect Biol. 2019 Jan 2;11(1):a032615. doi: 10.1101/cshperspect.a032615. Cold Spring Harb Perspect Biol. 2019. PMID: 29903714 Free PMC article. Review.
Cited by
-
Two Ways To Convert a Low-Affinity Potassium Channel to High Affinity: Control of Bacillus subtilis KtrCD by Glutamate.J Bacteriol. 2020 May 27;202(12):e00138-20. doi: 10.1128/JB.00138-20. Print 2020 May 27. J Bacteriol. 2020. PMID: 32253343 Free PMC article.
-
Effect of Nitrogen Concentration on the Biosynthesis of Citric Acid, Protein, and Lipids in the Yeast Yarrowia lipolytica.Biomolecules. 2022 Oct 4;12(10):1421. doi: 10.3390/biom12101421. Biomolecules. 2022. PMID: 36291630 Free PMC article.
-
Regulation of potassium uptake in Caulobacter crescentus.J Bacteriol. 2024 Sep 19;206(9):e0010724. doi: 10.1128/jb.00107-24. Epub 2024 Aug 12. J Bacteriol. 2024. PMID: 39133005
-
Monovalent metal ion binding promotes the first transesterification reaction in the spliceosome.Nat Commun. 2023 Dec 20;14(1):8482. doi: 10.1038/s41467-023-44174-2. Nat Commun. 2023. PMID: 38123540 Free PMC article.
-
The Peptidyl Transferase Center: a Window to the Past.Microbiol Mol Biol Rev. 2021 Dec 15;85(4):e0010421. doi: 10.1128/MMBR.00104-21. Epub 2021 Nov 10. Microbiol Mol Biol Rev. 2021. PMID: 34756086 Free PMC article.
References
-
- Sigel, A., Sigel, H. & Sigel, R. K. Structural and catalytic roles of metal ions in RNA. Met. Ions Life Sci. 9, vii–ix (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ANR-10-LABX-0030-INRT/Agence Nationale de la Recherche (French National Research Agency)/International
- ANR-15-CE11-0021-01/Agence Nationale de la Recherche (French National Research Agency)/International
- ANR-16-CE11-0007-01/Agence Nationale de la Recherche (French National Research Agency)/International
- DBF20160635745/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)/International
LinkOut - more resources
Full Text Sources
Medical

