Understanding heterogeneous tumor microenvironment in metastatic melanoma
- PMID: 31166985
- PMCID: PMC6550385
- DOI: 10.1371/journal.pone.0216485
Understanding heterogeneous tumor microenvironment in metastatic melanoma
Abstract
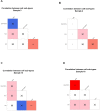
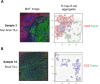
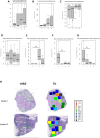
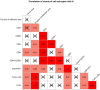
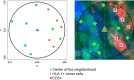
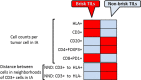
A systemic analysis of the tumor-immune interactions within the heterogeneous tumor microenvironment is of particular importance for understanding the antitumor immune response. We used multiplexed immunofluorescence to elucidate cellular spatial interactions and T-cell infiltrations in metastatic melanoma tumor microenvironment. We developed two novel computational approaches that enable infiltration clustering and single cell analysis-cell aggregate algorithm and cell neighborhood analysis algorithm-to reveal and to compare the spatial distribution of various immune cells relative to tumor cell in sub-anatomic tumor microenvironment areas. We showed that the heterogeneous tumor human leukocyte antigen-1 expressions differently affect the magnitude of cytotoxic T-cell infiltration and the distributions of CD20+ B cells and CD4+FOXP3+ regulatory T cells within and outside of T-cell infiltrated tumor areas. In a cohort of 166 stage III melanoma samples, high tumor human leukocyte antigen-1 expression is required but not sufficient for high T-cell infiltration, with significantly improved overall survival. Our results demonstrate that tumor cells with heterogeneous properties are associated with differential but predictable distributions of immune cells within heterogeneous tumor microenvironment with various biological features and impacts on clinical outcomes. It establishes tools necessary for systematic analysis of the tumor microenvironment, allowing the elucidation of the "homogeneous patterns" within the heterogeneous tumor microenvironment.
Conflict of interest statement
Authors AS, AS-P, CC, RZ, FG are currently employed at GE Global Research. This does not alter our adherence to PLOS ONE policies on sharing data and materials. Considering actual contribution to algorithm development which is the main novelty of this study, we added a line stating that two first authors contributed equally to this work.
Figures


Similar articles
-
T cells in the human metastatic melanoma microenvironment express site-specific homing receptors and retention integrins.Int J Cancer. 2014 Feb 1;134(3):563-74. doi: 10.1002/ijc.28391. Int J Cancer. 2014. PMID: 23873187 Free PMC article.
-
Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma.J Clin Invest. 2016 Sep 1;126(9):3447-52. doi: 10.1172/JCI87324. Epub 2016 Aug 15. J Clin Invest. 2016. PMID: 27525433 Free PMC article.
-
Immune cells in the melanoma microenvironment hold information for prediction of the risk of recurrence and response to treatment.Expert Rev Mol Diagn. 2014 Jul;14(6):643-6. doi: 10.1586/14737159.2014.928206. Epub 2014 Jun 10. Expert Rev Mol Diagn. 2014. PMID: 24914691
-
Modulation by MHC class I antigens of the biology of melanoma cells. Non-immunological mechanisms.Melanoma Res. 1993 Aug;3(4):285-9. Melanoma Res. 1993. PMID: 8219762 Review.
-
T-cell programming in pancreatic adenocarcinoma: a review.Cancer Gene Ther. 2017 Mar;24(3):106-113. doi: 10.1038/cgt.2016.66. Epub 2016 Dec 2. Cancer Gene Ther. 2017. PMID: 27910859 Review.
Cited by
-
Modulation of Lymphocyte Functions in the Microenvironment by Tumor Oncogenic Pathways.Front Immunol. 2022 May 19;13:883639. doi: 10.3389/fimmu.2022.883639. eCollection 2022. Front Immunol. 2022. PMID: 35663987 Free PMC article. Review.
-
Integrative analysis of bulk and single-cell RNA-seq reveals the molecular characterization of the immune microenvironment and oxidative stress signature in melanoma.Heliyon. 2024 Mar 15;10(6):e28244. doi: 10.1016/j.heliyon.2024.e28244. eCollection 2024 Mar 30. Heliyon. 2024. PMID: 38560689 Free PMC article.
-
ATF-3 expression inhibits melanoma growth by downregulating ERK and AKT pathways.Lab Invest. 2021 May;101(5):636-647. doi: 10.1038/s41374-020-00516-y. Epub 2020 Dec 9. Lab Invest. 2021. PMID: 33299127 Free PMC article.
-
Using the R Package Spatstat to Assess Inhibitory Effects of Microregional Hypoxia on the Infiltration of Cancers of the Head and Neck Region by Cytotoxic T Lymphocytes.Cancers (Basel). 2021 Apr 16;13(8):1924. doi: 10.3390/cancers13081924. Cancers (Basel). 2021. PMID: 33923522 Free PMC article.
-
From bench to bedside: Single-cell analysis for cancer immunotherapy.Cancer Cell. 2021 Aug 9;39(8):1062-1080. doi: 10.1016/j.ccell.2021.07.004. Epub 2021 Jul 29. Cancer Cell. 2021. PMID: 34329587 Free PMC article. Review.
References
-
- Robert C, Ribas A, Wolchok JD, Hodi FS, Hamid O, Kefford R, et al. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: a randomised dose-comparison cohort of a phase 1 trial. Lancet. 2014;384(9948):1109–1117. 10.1016/S0140-6736(14)60958-2 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

