In-depth Analysis of the Lid Subunits Assembly Mechanism in Mammals
- PMID: 31159305
- PMCID: PMC6627463
- DOI: 10.3390/biom9060213
In-depth Analysis of the Lid Subunits Assembly Mechanism in Mammals
Abstract
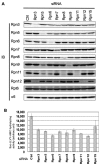
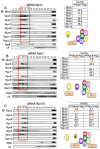
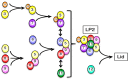
The 26S proteasome is a key player in the degradation of ubiquitinated proteins, comprising a 20S core particle (CP) and a 19S regulatory particle (RP). The RP is further divided into base and lid subcomplexes, which are assembled independently from each other. We have previously demonstrated the assembly pathway of the CP and the base by observing assembly intermediates resulting from knockdowns of each proteasome subunit and the assembly chaperones. In this study, we examine the assembly pathway of the mammalian lid, which remains to be elucidated. We show that the lid assembly pathway is conserved between humans and yeast. The final step is the incorporation of Rpn12 into the assembly intermediate consisting of two modular complexes, Rpn3-7-15 and Rpn5-6-8-9-11, in both humans and yeast. Furthermore, we dissect the assembly pathways of the two modular complexes by the knockdown of each lid subunit.
Keywords: 19S regulatory particle; 26S proteasome; Rpn proteins; assembly; lid subcomplex.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures






Similar articles
-
Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining.Mol Cell. 2011 Dec 23;44(6):907-17. doi: 10.1016/j.molcel.2011.11.020. Mol Cell. 2011. PMID: 22195964 Free PMC article.
-
Base-CP proteasome can serve as a platform for stepwise lid formation.Biosci Rep. 2015 Jan 27;35(3):e00194. doi: 10.1042/BSR20140173. Biosci Rep. 2015. PMID: 26182356 Free PMC article.
-
Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae.Biochem Biophys Res Commun. 2010 Jun 11;396(4):1048-53. doi: 10.1016/j.bbrc.2010.05.061. Epub 2010 May 21. Biochem Biophys Res Commun. 2010. PMID: 20471955
-
Assembly manual for the proteasome regulatory particle: the first draft.Biochem Soc Trans. 2010 Feb;38(Pt 1):6-13. doi: 10.1042/BST0380006. Biochem Soc Trans. 2010. PMID: 20074027 Free PMC article. Review.
-
Proteasome assembly.Cell Mol Life Sci. 2014 Dec;71(24):4729-45. doi: 10.1007/s00018-014-1699-8. Epub 2014 Aug 9. Cell Mol Life Sci. 2014. PMID: 25107634 Free PMC article. Review.
Cited by
-
Proteasomal subunit depletions differentially affect germline integrity in C. elegans.Front Cell Dev Biol. 2022 Aug 17;10:901320. doi: 10.3389/fcell.2022.901320. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36060813 Free PMC article.
-
Structural dynamics of the human COP9 signalosome revealed by cross-linking mass spectrometry and integrative modeling.Proc Natl Acad Sci U S A. 2020 Feb 25;117(8):4088-4098. doi: 10.1073/pnas.1915542117. Epub 2020 Feb 7. Proc Natl Acad Sci U S A. 2020. PMID: 32034103 Free PMC article.
-
Hepatitis B virus infection disrupts homologous recombination in hepatocellular carcinoma by stabilizing resection inhibitor ADRM1.J Clin Invest. 2023 Dec 1;133(23):e171533. doi: 10.1172/JCI171533. J Clin Invest. 2023. PMID: 37815873 Free PMC article.
-
The RPN12a proteasome subunit is essential for the multiple hormonal homeostasis controlling the progression of leaf senescence.Commun Biol. 2022 Sep 30;5(1):1043. doi: 10.1038/s42003-022-03998-2. Commun Biol. 2022. PMID: 36180574 Free PMC article.
-
Protective Roles of Cytosolic and Plastidal Proteasomes on Abiotic Stress and Pathogen Invasion.Plants (Basel). 2020 Jul 2;9(7):832. doi: 10.3390/plants9070832. Plants (Basel). 2020. PMID: 32630761 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

