Probe dependency in the determination of ligand binding kinetics at a prototypical G protein-coupled receptor
- PMID: 31133718
- PMCID: PMC6536503
- DOI: 10.1038/s41598-019-44025-5
Probe dependency in the determination of ligand binding kinetics at a prototypical G protein-coupled receptor
Abstract
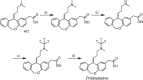
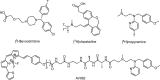
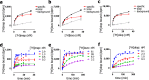
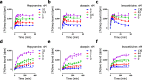
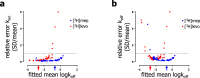
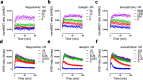
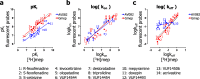
Drug-target binding kinetics are suggested to be important parameters for the prediction of in vivo drug-efficacy. For G protein-coupled receptors (GPCRs), the binding kinetics of ligands are typically determined using association binding experiments in competition with radiolabelled probes, followed by analysis with the widely used competitive binding kinetics theory developed by Motulsky and Mahan. Despite this, the influence of the radioligand binding kinetics on the kinetic parameters derived for the ligands tested is often overlooked. To address this, binding rate constants for a series of histamine H1 receptor (H1R) antagonists were determined using radioligands with either slow (low koff) or fast (high koff) dissociation characteristics. A correlation was observed between the probe-specific datasets for the kinetic binding affinities, association rate constants and dissociation rate constants. However, the magnitude and accuracy of the binding rate constant-values was highly dependent on the used radioligand probe. Further analysis using recently developed fluorescent binding methods corroborates the finding that the Motulsky-Mahan methodology is limited by the employed assay conditions. The presented data suggest that kinetic parameters of GPCR ligands depend largely on the characteristics of the probe used and results should therefore be viewed within the experimental context and limitations of the applied methodology.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
BRET-based β-arrestin2 recruitment to the histamine H1 receptor for investigating antihistamine binding kinetics.Pharmacol Res. 2016 Sep;111:679-687. doi: 10.1016/j.phrs.2016.07.034. Epub 2016 Jul 26. Pharmacol Res. 2016. PMID: 27468652
-
Analytical method for simultaneously measuring ex vivo drug receptor occupancy and dissociation rate: application to (R)-dimethindene occupancy of central histamine H1 receptors.J Recept Signal Transduct Res. 2009;29(2):84-93. doi: 10.1080/10799890902721339. J Recept Signal Transduct Res. 2009. PMID: 19308787
-
Influence of the N-terminus and the E2-loop onto the binding kinetics of the antagonist mepyramine and the partial agonist phenoprodifen to H(1)R.Biochem Pharmacol. 2011 Dec 15;82(12):1910-8. doi: 10.1016/j.bcp.2011.09.005. Epub 2011 Sep 16. Biochem Pharmacol. 2011. PMID: 21933664
-
Reversible and irreversible labelling of H1- and H2 -receptors using novel [125I] probes.Agents Actions Suppl. 1991;33:123-44. doi: 10.1007/978-3-0348-7309-3_10. Agents Actions Suppl. 1991. PMID: 1675831 Review.
-
Delineation of receptor-ligand interactions at the human histamine H1 receptor by a combined approach of site-directed mutagenesis and computational techniques - or - how to bind the H1 receptor.Arch Pharm (Weinheim). 2005 Jun;338(5-6):248-59. doi: 10.1002/ardp.200400998. Arch Pharm (Weinheim). 2005. PMID: 15952243 Review.
Cited by
-
A structure-kinetic relationship study using matched molecular pair analysis.RSC Med Chem. 2020 Sep 21;11(11):1285-1294. doi: 10.1039/d0md00178c. RSC Med Chem. 2020. PMID: 34085042 Free PMC article.
-
The Impact of the Secondary Binding Pocket on the Pharmacology of Class A GPCRs.Front Pharmacol. 2022 Mar 9;13:847788. doi: 10.3389/fphar.2022.847788. eCollection 2022. Front Pharmacol. 2022. PMID: 35355719 Free PMC article. Review.
-
Perspective: Implications of Ligand-Receptor Binding Kinetics for Therapeutic Targeting of G Protein-Coupled Receptors.ACS Pharmacol Transl Sci. 2020 Mar 18;3(2):179-189. doi: 10.1021/acsptsci.0c00012. eCollection 2020 Apr 10. ACS Pharmacol Transl Sci. 2020. PMID: 32296761 Free PMC article. Review.
-
Exploring the Effect of Cyclization of Histamine H1 Receptor Antagonists on Ligand Binding Kinetics.ACS Omega. 2021 May 7;6(19):12755-12768. doi: 10.1021/acsomega.0c06358. eCollection 2021 May 18. ACS Omega. 2021. PMID: 34056427 Free PMC article.
-
CLAffinity: A Software Tool for Identification of Optimum Ligand Affinity for Competition-Based Primary Screens.J Chem Inf Model. 2022 May 23;62(10):2264-2268. doi: 10.1021/acs.jcim.2c00285. Epub 2022 Apr 20. J Chem Inf Model. 2022. PMID: 35442032 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

