PatchSearch: a web server for off-target protein identification
- PMID: 31131411
- PMCID: PMC6602448
- DOI: 10.1093/nar/gkz478
PatchSearch: a web server for off-target protein identification
Abstract
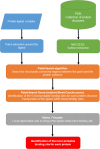
The large number of proteins found in the human body implies that a drug may interact with many proteins, called off-target proteins, besides its intended target. The PatchSearch web server provides an automated workflow that allows users to identify structurally conserved binding sites at the protein surfaces in a set of user-supplied protein structures. Thus, this web server may help to detect potential off-target protein. It takes as input a protein complexed with a ligand and identifies within user-defined or predefined collections of protein structures, those having a binding site compatible with this ligand in terms of geometry and physicochemical properties. It is based on a non-sequential local alignment of the patch over the entire protein surface. Then the PatchSearch web server proposes a ligand binding mode for the potential off-target, as well as an estimated affinity calculated by the Vinardo scoring function. This novel tool is able to efficiently detects potential interactions of ligands with distant off-target proteins. Furthermore, by facilitating the discovery of unexpected off-targets, PatchSearch could contribute to the repurposing of existing drugs. The server is freely available at http://bioserv.rpbs.univ-paris-diderot.fr/services/PatchSearch.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

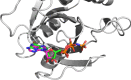
Similar articles
-
SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules.Nucleic Acids Res. 2019 Jul 2;47(W1):W357-W364. doi: 10.1093/nar/gkz382. Nucleic Acids Res. 2019. PMID: 31106366 Free PMC article.
-
Drug ReposER: a web server for predicting similar amino acid arrangements to known drug binding interfaces for potential drug repositioning.Nucleic Acids Res. 2019 Jul 2;47(W1):W350-W356. doi: 10.1093/nar/gkz391. Nucleic Acids Res. 2019. PMID: 31106379 Free PMC article.
-
MTiOpenScreen: a web server for structure-based virtual screening.Nucleic Acids Res. 2015 Jul 1;43(W1):W448-54. doi: 10.1093/nar/gkv306. Epub 2015 Apr 8. Nucleic Acids Res. 2015. PMID: 25855812 Free PMC article.
-
NMR studies of ligand binding.Curr Opin Struct Biol. 2018 Feb;48:16-22. doi: 10.1016/j.sbi.2017.09.001. Epub 2017 Oct 7. Curr Opin Struct Biol. 2018. PMID: 29017071 Review.
-
NMR in structure-based drug design.Essays Biochem. 2017 Nov 8;61(5):485-493. doi: 10.1042/EBC20170037. Print 2017 Nov 8. Essays Biochem. 2017. PMID: 29118095 Review.
Cited by
-
LigTMap: ligand and structure-based target identification and activity prediction for small molecular compounds.J Cheminform. 2021 Jun 10;13(1):44. doi: 10.1186/s13321-021-00523-1. J Cheminform. 2021. PMID: 34112240 Free PMC article.
-
VirtuousPocketome: a computational tool for screening protein-ligand complexes to identify similar binding sites.Sci Rep. 2024 Mar 15;14(1):6296. doi: 10.1038/s41598-024-56893-7. Sci Rep. 2024. PMID: 38491261 Free PMC article.
References
-
- Vulpetti A., Kalliokoski T., Milletti F.. Chemogenomics in drug discovery: computational methods based on the comparison of binding sites. Future Med. Chem. 2012; 4:1971–1979. - PubMed
-
- Jalencas X., Mestres J.. Identification of similar binding sites to detect distant polypharmacology. Mol. Inform. 2013; 32:976–990. - PubMed
-
- Ritchie D.W., Kemp G.J.L.. Fast computation, rotation and comparison of low resolution spherical harmonic molecular surfaces. J. Comput. Chem. 1999; 20:383–395.
-
- Morris R.J., Najmanovich R.J., Kahraman A., Thornton J.M.. Real spherical harmonic expansion coefficients as 3D shape descriptors for protein binding pocket and ligand comparisons. Bioinformatics. 2005; 21:2347–2355. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

