Conservation of molecular and cellular phenotypes of invariant NKT cells between humans and non-human primates
- PMID: 31123763
- PMCID: PMC6647187
- DOI: 10.1007/s00251-019-01118-9
Conservation of molecular and cellular phenotypes of invariant NKT cells between humans and non-human primates
Abstract
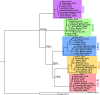
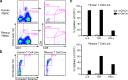
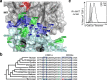
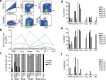
Invariant NKT (iNKT) cells in both humans and non-human primates are activated by the glycolipid antigen, α-galactosylceramide (α-GalCer). However, the extent to which the molecular mechanisms of antigen recognition and in vivo phenotypes of iNKT cells are conserved among primate species has not been determined. Using an evolutionary genetic approach, we found a lack of diversifying selection in CD1 genes over 45 million years of evolution, which stands in stark contrast to the history of the MHC system for presenting peptide antigens to T cells. The invariant T cell receptor (TCR)-α chain was strictly conserved across all seven primate clades. Invariant NKT cells from rhesus macaques (Macaca mulatta) bind human CD1D-α-GalCer tetramer and are activated by α-GalCer-loaded human CD1D transfectants. The dominant TCR-β chain cloned from a rhesus-derived iNKT cell line is nearly identical to that found in the human iNKT TCR, and transduction of the rhesus iNKT TCR into human Jurkat cells show that it is sufficient for binding human CD1D-α-GalCer tetramer. Finally, we used a 20-color flow cytometry panel to probe tissue phenotypes of iNKT cells in a cohort of rhesus macaques. We discovered several tissue-resident iNKT populations that have not been previously described in non-human primates but are known in humans, such as TCR-γδ iNKTs. These data reveal a diversity of iNKT cell phenotypes despite convergent evolution of the genes required for lipid antigen presentation and recognition in humans and non-human primates.
Keywords: CD1D; Non-human primate; T cell receptor; iNKT cells.
Figures
Similar articles
-
The complementarity determining region 2 of BV8S2 (V beta 8.2) contributes to antigen recognition by rat invariant NKT cell TCR.J Immunol. 2006 Jun 15;176(12):7447-55. doi: 10.4049/jimmunol.176.12.7447. J Immunol. 2006. PMID: 16751390
-
The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation.J Exp Med. 2007 May 14;204(5):1131-44. doi: 10.1084/jem.20062342. Epub 2007 May 7. J Exp Med. 2007. PMID: 17485514 Free PMC article.
-
The CD1d natural killer T-cell antigen presentation pathway is highly conserved between humans and rhesus macaques.Immunogenetics. 2003 Feb;54(11):776-81. doi: 10.1007/s00251-002-0527-8. Epub 2003 Feb 6. Immunogenetics. 2003. PMID: 12618910
-
Role of invariant natural killer T (iNKT) cells in systemic lupus erythematosus.Curr Med Chem. 2008;15(18):1778-87. doi: 10.2174/092986708785132988. Curr Med Chem. 2008. PMID: 18691038 Review.
-
Invariant NKT and NKT-like lymphocytes: two different T cell subsets that are differentially affected by ageing.Exp Gerontol. 2007 Aug;42(8):703-8. doi: 10.1016/j.exger.2007.05.002. Epub 2007 May 21. Exp Gerontol. 2007. PMID: 17604928 Review.
Cited by
-
A simple assay to quantify mycobacterial lipid antigen-specific T cell receptors in human tissues and blood.PLoS Negl Trop Dis. 2021 Dec 16;15(12):e0010018. doi: 10.1371/journal.pntd.0010018. eCollection 2021 Dec. PLoS Negl Trop Dis. 2021. PMID: 34914694 Free PMC article.
-
Modulation of Immune Responses to Influenza A Virus Vaccines by Natural Killer T Cells.Front Immunol. 2020 Oct 20;11:2172. doi: 10.3389/fimmu.2020.02172. eCollection 2020. Front Immunol. 2020. PMID: 33193296 Free PMC article. Review.
-
Functional and biological implications of clonotypic diversity among human donor-unrestricted T cells.Immunol Cell Biol. 2024 Jul;102(6):474-486. doi: 10.1111/imcb.12751. Epub 2024 Apr 24. Immunol Cell Biol. 2024. PMID: 38659280 Review.
-
The role of donor-unrestricted T-cells, innate lymphoid cells, and NK cells in anti-mycobacterial immunity.Immunol Rev. 2021 May;301(1):30-47. doi: 10.1111/imr.12948. Epub 2021 Feb 2. Immunol Rev. 2021. PMID: 33529407 Free PMC article. Review.
-
Targeting adrenergic receptors to mitigate invariant natural killer T cells-induced acute liver injury.iScience. 2023 Sep 16;26(10):107947. doi: 10.1016/j.isci.2023.107947. eCollection 2023 Oct 20. iScience. 2023. PMID: 37841583 Free PMC article.
References
-
- Balk SP, Bleicher PA, Terhorst C. Isolation and expression of cDNA encoding the murine homologues of CD1. J Immunol. 1991;146:768–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

