SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules
- PMID: 31106366
- PMCID: PMC6602486
- DOI: 10.1093/nar/gkz382
SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules
Abstract
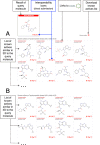
SwissTargetPrediction is a web tool, on-line since 2014, that aims to predict the most probable protein targets of small molecules. Predictions are based on the similarity principle, through reverse screening. Here, we describe the 2019 version, which represents a major update in terms of underlying data, backend and web interface. The bioactivity data were updated, the model retrained and similarity thresholds redefined. In the new version, the predictions are performed by searching for similar molecules, in 2D and 3D, within a larger collection of 376 342 compounds known to be experimentally active on an extended set of 3068 macromolecular targets. An efficient backend implementation allows to speed up the process that returns results for a druglike molecule on human proteins in 15-20 s. The refreshed web interface enhances user experience with new features for easy input and improved analysis. Interoperability capacity enables straightforward submission of any input or output molecule to other on-line computer-aided drug design tools, developed by the SIB Swiss Institute of Bioinformatics. High levels of predictive performance were maintained despite more extended biological and chemical spaces to be explored, e.g. achieving at least one correct human target in the top 15 predictions for >70% of external compounds. The new SwissTargetPrediction is available free of charge (www.swisstargetprediction.ch).
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
PatchSearch: a web server for off-target protein identification.Nucleic Acids Res. 2019 Jul 2;47(W1):W365-W372. doi: 10.1093/nar/gkz478. Nucleic Acids Res. 2019. PMID: 31131411 Free PMC article.
-
SwissTargetPrediction: a web server for target prediction of bioactive small molecules.Nucleic Acids Res. 2014 Jul;42(Web Server issue):W32-8. doi: 10.1093/nar/gku293. Epub 2014 May 3. Nucleic Acids Res. 2014. PMID: 24792161 Free PMC article.
-
SwissSimilarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening.J Chem Inf Model. 2016 Aug 22;56(8):1399-404. doi: 10.1021/acs.jcim.6b00174. Epub 2016 Jul 19. J Chem Inf Model. 2016. PMID: 27391578
-
Application of the SwissDrugDesign Online Resources in Virtual Screening.Int J Mol Sci. 2019 Sep 18;20(18):4612. doi: 10.3390/ijms20184612. Int J Mol Sci. 2019. PMID: 31540350 Free PMC article. Review.
-
The SGC beyond structural genomics: redefining the role of 3D structures by coupling genomic stratification with fragment-based discovery.Essays Biochem. 2017 Nov 8;61(5):495-503. doi: 10.1042/EBC20170051. Print 2017 Nov 8. Essays Biochem. 2017. PMID: 29118096 Free PMC article. Review.
Cited by
-
Network Pharmacology, Molecular Dynamics and In Vitro Assessments of Indigenous Herbal Formulations for Alzheimer's Therapy.Life (Basel). 2024 Sep 25;14(10):1222. doi: 10.3390/life14101222. Life (Basel). 2024. PMID: 39459522 Free PMC article.
-
Integrated network pharmacology and experimental analysis unveil multi-targeted effect of 18α- glycyrrhetinic acid against non-small cell lung cancer.Front Pharmacol. 2022 Oct 12;13:1018974. doi: 10.3389/fphar.2022.1018974. eCollection 2022. Front Pharmacol. 2022. PMID: 36313358 Free PMC article.
-
LC-MS/MS Phytochemical Profiling, Antioxidant Activity, and Cytotoxicity of the Ethanolic Extract of Atriplex halimus L. against Breast Cancer Cell Lines: Computational Studies and Experimental Validation.Pharmaceuticals (Basel). 2022 Sep 16;15(9):1156. doi: 10.3390/ph15091156. Pharmaceuticals (Basel). 2022. PMID: 36145377 Free PMC article.
-
Astragaloside IV ameliorates pressure overload-induced heart failure by enhancing angiogenesis through HSF1/VEGF pathway.Heliyon. 2024 Sep 1;10(17):e37019. doi: 10.1016/j.heliyon.2024.e37019. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39296120 Free PMC article.
-
Exploring the mechanism of andrographolide in the treatment of gastric cancer through network pharmacology and molecular docking.Sci Rep. 2022 Nov 1;12(1):18413. doi: 10.1038/s41598-022-18319-0. Sci Rep. 2022. PMID: 36319798 Free PMC article.
References
-
- Byrne R., Schneider G.. In silico target prediction for small molecules. Methods Mol. Biol. 2019; 1888:273–309. - PubMed
-
- Lavecchia A., Cerchia C.. In silico methods to address polypharmacology: current status, applications and future perspectives. Drug Discov. Today. 2016; 21:288–298. - PubMed
-
- Cereto-Massagué A., Ojeda M.J., Valls C., Mulero M., Pujadas G., Garcia-Vallvé S.. Tools for in silico target fishing. Methods. 2015; 71:98–103. - PubMed
-
- Ding H., Takigawa I., Mamitsuka H., Zhu S.. Similarity-based machine learning methods for predicting drug-target interactions: a brief review. Brief. Bioinformatics. 2014; 15:734–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

