Pheromones and Nutritional Signals Regulate the Developmental Reliance on let-7 Family MicroRNAs in C. elegans
- PMID: 31104929
- PMCID: PMC7245018
- DOI: 10.1016/j.cub.2019.04.034
Pheromones and Nutritional Signals Regulate the Developmental Reliance on let-7 Family MicroRNAs in C. elegans
Abstract
Adverse environmental conditions can affect rates of animal developmental progression and lead to temporary developmental quiescence (diapause), exemplified by the dauer larva stage of the nematode Caenorhabditis elegans (C. elegans). Remarkably, patterns of cell division and temporal cell-fate progression in C. elegans larvae are not affected by changes in developmental trajectory. However, the underlying physiological and gene regulatory mechanisms that ensure robust developmental patterning despite substantial plasticity in developmental progression are largely unknown. Here, we report that diapause-inducing pheromones correct heterochronic developmental cell lineage defects caused by insufficient expression of let-7 family microRNAs in C. elegans. Moreover, two conserved endocrine signaling pathways, DAF-7/TGF-β and DAF-2/Insulin, that confer on the larva diapause and non-diapause alternative developmental trajectories interact with the nuclear hormone receptor, DAF-12, to initiate and regulate a rewiring of the genetic circuitry controlling temporal cell fates. This rewiring includes engagement of certain heterochronic genes, lin-46, lin-4, and nhl-2, that are previously associated with an altered genetic program in post-diapause animals, in combination with a novel ligand-independent DAF-12 activity, to downregulate the critical let-7 family target Hunchback-like-1 (HBL-1). Our results show how pheromone or endocrine signaling pathways can coordinately regulate both developmental progression and cell-fate transitions in C. elegans larvae under stress so that the developmental schedule of cell fates remains unaffected by changes in developmental trajectory.
Keywords: ascarosides; dauer larva; developmental robustness; endocrine signaling; heterochronic genes; let-7; microRNAs; pheromones; reprogramming; stem cell.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures
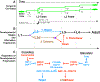



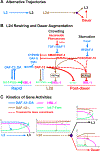
Comment in
-
Developmental Timing: Honey, I Reprogrammed the Kids.Curr Biol. 2019 Jun 3;29(11):R420-R422. doi: 10.1016/j.cub.2019.04.054. Curr Biol. 2019. PMID: 31163147
Similar articles
-
Dauer larva quiescence alters the circuitry of microRNA pathways regulating cell fate progression in C. elegans.Development. 2012 Jun;139(12):2177-86. doi: 10.1242/dev.075986. Development. 2012. PMID: 22619389 Free PMC article.
-
A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans.Proc Natl Acad Sci U S A. 2009 Nov 3;106(44):18668-73. doi: 10.1073/pnas.0908131106. Epub 2009 Oct 14. Proc Natl Acad Sci U S A. 2009. PMID: 19828440 Free PMC article.
-
acn-1, a C. elegans homologue of ACE, genetically interacts with the let-7 microRNA and other heterochronic genes.Cell Cycle. 2017 Oct 2;16(19):1800-1809. doi: 10.1080/15384101.2017.1344798. Epub 2017 Sep 21. Cell Cycle. 2017. PMID: 28933985 Free PMC article.
-
[Genetics and evolution of developmental plasticity in the nematode C. elegans: Environmental induction of the dauer stage].Biol Aujourdhui. 2020;214(1-2):45-53. doi: 10.1051/jbio/2020006. Epub 2020 Aug 10. Biol Aujourdhui. 2020. PMID: 32773029 Review. French.
-
Developmental transitions in C. elegans larval stages.Curr Top Dev Biol. 2013;105:153-80. doi: 10.1016/B978-0-12-396968-2.00006-3. Curr Top Dev Biol. 2013. PMID: 23962842 Review.
Cited by
-
Nematode Pheromones: Structures and Functions.Molecules. 2023 Mar 6;28(5):2409. doi: 10.3390/molecules28052409. Molecules. 2023. PMID: 36903652 Free PMC article. Review.
-
Ascaroside Pheromones: Chemical Biology and Pleiotropic Neuronal Functions.Int J Mol Sci. 2019 Aug 9;20(16):3898. doi: 10.3390/ijms20163898. Int J Mol Sci. 2019. PMID: 31405082 Free PMC article. Review.
-
C. elegans LIN-28 controls temporal cell fate progression by regulating LIN-46 expression via the 5' UTR of lin-46 mRNA.Cell Rep. 2021 Sep 7;36(10):109670. doi: 10.1016/j.celrep.2021.109670. Cell Rep. 2021. PMID: 34496246 Free PMC article.
-
A life cycle alteration can correct molting defects in Caenorhabditis elegans.Dev Biol. 2022 Mar;483:143-156. doi: 10.1016/j.ydbio.2022.01.001. Epub 2022 Jan 15. Dev Biol. 2022. PMID: 35038442 Free PMC article.
-
Recent advances in understanding microRNA function and regulation in C. elegans.Semin Cell Dev Biol. 2024 Feb 15;154(Pt A):4-13. doi: 10.1016/j.semcdb.2023.03.011. Epub 2023 Apr 11. Semin Cell Dev Biol. 2024. PMID: 37055330 Review.
References
-
- Sulston JE, and Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol 56, 110–156. - PubMed
-
- Ambros V, and Horvitz HR (1984). Heterochronic mutants of the nematode Caenorhabditis elegans. Science 226, 409–16. - PubMed
-
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, and Slack FJ (2003). The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable MicroRNA target. Dev. Cell 4, 639–650. - PubMed
-
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, and Rougvie AE (2003). The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4, 625–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

