The processivity factor Pol32 mediates nuclear localization of DNA polymerase delta and prevents chromosomal fragile site formation in Drosophila development
- PMID: 31100062
- PMCID: PMC6542543
- DOI: 10.1371/journal.pgen.1008169
The processivity factor Pol32 mediates nuclear localization of DNA polymerase delta and prevents chromosomal fragile site formation in Drosophila development
Abstract
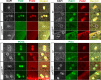
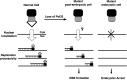
The Pol32 protein is one of the universal subunits of DNA polymerase δ (Pol δ), which is responsible for genome replication in eukaryotic cells. Although the role of Pol32 in DNA repair has been well-characterized, its exact function in genome replication remains obscure as studies in single cell systems have not established an essential role for Pol32 in the process. Here we characterize Pol32 in the context of Drosophila melanogaster development. In the rapidly dividing embryonic cells, loss of Pol32 halts genome replication as it specifically disrupts Pol δ localization to the nucleus. This function of Pol32 in facilitating the nuclear import of Pol δ would be similar to that of accessory subunits of DNA polymerases from mammalian Herpes viruses. In post-embryonic cells, loss of Pol32 reveals mitotic fragile sites in the Drosophila genome, a defect more consistent with Pol32's role as a polymerase processivity factor. Interestingly, these fragile sites do not favor repetitive sequences in heterochromatin, with the rDNA locus being a striking exception. Our study uncovers a possibly universal function for DNA polymerase ancillary factors and establishes a powerful system for the study of chromosomal fragile sites in a non-mammalian organism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Loss of Pol32 in Drosophila melanogaster causes chromosome instability and suppresses variegation.PLoS One. 2015 Mar 31;10(3):e0120859. doi: 10.1371/journal.pone.0120859. eCollection 2015. PLoS One. 2015. PMID: 25826374 Free PMC article.
-
A novel variant of DNA polymerase ζ, Rev3ΔC, highlights differential regulation of Pol32 as a subunit of polymerase δ versus ζ in Saccharomyces cerevisiae.DNA Repair (Amst). 2014 Dec;24:138-149. doi: 10.1016/j.dnarep.2014.04.013. Epub 2014 May 10. DNA Repair (Amst). 2014. PMID: 24819597 Free PMC article.
-
Translesion polymerase eta both facilitates DNA replication and promotes increased human genetic variation at common fragile sites.Proc Natl Acad Sci U S A. 2021 Nov 30;118(48):e2106477118. doi: 10.1073/pnas.2106477118. Proc Natl Acad Sci U S A. 2021. PMID: 34815340 Free PMC article.
-
Structure and function of eukaryotic DNA polymerase δ.Subcell Biochem. 2012;62:217-36. doi: 10.1007/978-94-007-4572-8_12. Subcell Biochem. 2012. PMID: 22918588 Review.
-
DNA polymerase delta, an essential enzyme for DNA transactions.Biol Chem. 1997 May;378(5):345-62. doi: 10.1515/bchm.1997.378.5.345. Biol Chem. 1997. PMID: 9191022 Review.
Cited by
-
The DEAD-Box Protein Rok1 Coordinates Ribosomal RNA Processing in Association with Rrp5 in Drosophila.Int J Mol Sci. 2022 May 19;23(10):5685. doi: 10.3390/ijms23105685. Int J Mol Sci. 2022. PMID: 35628496 Free PMC article.
-
Genomic instability and cancer: lessons from Drosophila.Open Biol. 2020 Jun;10(6):200060. doi: 10.1098/rsob.200060. Epub 2020 Jun 3. Open Biol. 2020. PMID: 32485126 Free PMC article. Review.
-
Under the magnifying glass: The ups and downs of rDNA copy number.Semin Cell Dev Biol. 2023 Feb 28;136:38-48. doi: 10.1016/j.semcdb.2022.05.006. Epub 2022 May 18. Semin Cell Dev Biol. 2023. PMID: 35595601 Free PMC article. Review.
-
Association of Mutations in Replicative DNA Polymerase Genes with Human Disease: Possible Application of Drosophila Models for Studies.Int J Mol Sci. 2023 Apr 29;24(9):8078. doi: 10.3390/ijms24098078. Int J Mol Sci. 2023. PMID: 37175782 Free PMC article. Review.
-
The DNA polymerases of Drosophila melanogaster.Fly (Austin). 2020 Mar-Dec;14(1-4):49-61. doi: 10.1080/19336934.2019.1710076. Epub 2020 Jan 14. Fly (Austin). 2020. PMID: 31933406 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

