Glutamine to proline conversion is associated with response to glutaminase inhibition in breast cancer
- PMID: 31088535
- PMCID: PMC6518522
- DOI: 10.1186/s13058-019-1141-0
Glutamine to proline conversion is associated with response to glutaminase inhibition in breast cancer
Abstract
Introduction: Glutaminase inhibitors target cancer cells by blocking the conversion of glutamine to glutamate, thereby potentially interfering with anaplerosis and synthesis of amino acids and glutathione. The drug CB-839 has shown promising effects in preclinical experiments and is currently undergoing clinical trials in several human malignancies, including triple-negative breast cancer (TNBC). However, response to glutaminase inhibitors is variable and there is a need for identification of predictive response biomarkers. The aim of this study was to determine how glutamine is utilized in two patient-derived xenograft (PDX) models of breast cancer representing luminal-like/ER+ (MAS98.06) and basal-like/triple-negative (MAS98.12) breast cancer and to explore the metabolic effects of CB-839 treatment.
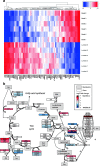
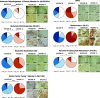
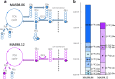
Experimental: MAS98.06 and MAS98.12 PDX mice received CB-839 (200 mg/kg) or drug vehicle two times daily p.o. for up to 28 days (n = 5 per group), and the effect on tumor growth was evaluated. Expression of 60 genes and seven glutaminolysis key enzymes were determined using gene expression microarray analysis and immunohistochemistry (IHC), respectively, in untreated tumors. Uptake and conversion of glutamine were determined in the PDX models using HR MAS MRS after i.v. infusion of [5-13C] glutamine when the models had received CB-839 (200 mg/kg) or vehicle for 2 days (n = 5 per group).
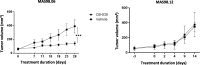
Results: Tumor growth measurements showed that CB-839 significantly inhibited tumor growth in MAS98.06 tumors, but not in MAS98.12 tumors. Gene expression and IHC analysis indicated a higher proline synthesis from glutamine in untreated MAS98.06 tumors. This was confirmed by HR MAS MRS of untreated tumors demonstrating that MAS98.06 used glutamine to produce proline, glutamate, and alanine, and MAS98.12 to produce glutamate and lactate. In both models, treatment with CB-839 resulted in accumulation of glutamine. In addition, CB-839 caused depletion of alanine, proline, and glutamate ([1-13C] glutamate) in the MAS98.06 model.
Conclusion: Our findings indicate that TNBCs may not be universally sensitive to glutaminase inhibitors. The major difference in the metabolic fate of glutamine between responding MAS98.06 xenografts and non-responding MAS98.12 xenografts is the utilization of glutamine for production of proline. We therefore suggest that addiction to proline synthesis from glutamine is associated with response to CB-839 in breast cancer. The effect of glutaminase inhibition in two breast cancer patient-derived xenograft (PDX) models. 13C HR MAS MRS analysis of tumor tissue from CB-839-treated and untreated models receiving 13C-labeled glutamine ([5-13C] Gln) shows that the glutaminase inhibitor CB-839 is causing an accumulation of glutamine (arrow up) in two PDX models representing luminal-like breast cancer (MAS98.06) and basal-like breast cancer (MAS98.12). In MAS98.06 tumors, CB-839 is in addition causing depletion of proline ([5-13C] Pro), alanine ([1-13C] Ala), and glutamate ([1-13C] Glu), which could explain why CB-839 causes tumor growth inhibition in MAS98.06 tumors, but not in MAS98.12 tumors.
Keywords: 13C MRS; Aldehyde dehydrogenase 18 family member A1 (ALDH18A1); CB-839; Cancer treatment; Gene expression analysis; Glutaminase; Glutaminase inhibitor; High-resolution magic angle spinning MR spectroscopy (HR MAS MRS); Immunohistochemistry; Patient-derived xenograft (PDX).
Conflict of interest statement
Ethics approval and consent to participate
All procedures and experiments involving animals were approved by the Norwegian Animal Research Authority (FOTS ID: 7713 and 9126) and carried out according to the European Convention for the Protection of Vertebrates used for Scientific Purposes.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

Similar articles
-
Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer.Mol Cancer Ther. 2014 Apr;13(4):890-901. doi: 10.1158/1535-7163.MCT-13-0870. Epub 2014 Feb 12. Mol Cancer Ther. 2014. PMID: 24523301
-
Glutamate-Weighted Chemical Exchange Saturation Transfer Magnetic Resonance Imaging Detects Glutaminase Inhibition in a Mouse Model of Triple-Negative Breast Cancer.Cancer Res. 2018 Oct 1;78(19):5521-5526. doi: 10.1158/0008-5472.CAN-17-3988. Epub 2018 Aug 2. Cancer Res. 2018. PMID: 30072394 Free PMC article.
-
13C high-resolution-magic angle spinning MRS reveals differences in glucose metabolism between two breast cancer xenograft models with different gene expression patterns.NMR Biomed. 2011 Dec;24(10):1243-52. doi: 10.1002/nbm.1683. Epub 2011 Apr 4. NMR Biomed. 2011. PMID: 21462378
-
Recent Development of Small Molecule Glutaminase Inhibitors.Curr Top Med Chem. 2018;18(6):432-443. doi: 10.2174/1568026618666180525100830. Curr Top Med Chem. 2018. PMID: 29793408 Review.
-
Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer.Semin Cell Dev Biol. 2020 Feb;98:34-43. doi: 10.1016/j.semcdb.2019.05.012. Epub 2019 May 22. Semin Cell Dev Biol. 2020. PMID: 31100352 Review.
Cited by
-
Reproducible Lipid Alterations in Patient-Derived Breast Cancer Xenograft FFPE Tissue Identified with MALDI MSI for Pre-Clinical and Clinical Application.Metabolites. 2021 Aug 26;11(9):577. doi: 10.3390/metabo11090577. Metabolites. 2021. PMID: 34564393 Free PMC article.
-
Targeting glutamine metabolism as a therapeutic strategy for cancer.Exp Mol Med. 2023 Apr;55(4):706-715. doi: 10.1038/s12276-023-00971-9. Epub 2023 Apr 3. Exp Mol Med. 2023. PMID: 37009798 Free PMC article. Review.
-
The Role of Pi, Glutamine and the Essential Amino Acids in Modulating the Metabolism in Diabetes and Cancer.J Diabetes Metab Disord. 2020 Aug 19;19(2):1731-1775. doi: 10.1007/s40200-020-00566-5. eCollection 2020 Dec. J Diabetes Metab Disord. 2020. PMID: 33520860 Free PMC article. Review.
-
Potential of Anti-Cancer Activity of Secondary Metabolic Products from Marine Fungi.J Fungi (Basel). 2021 May 30;7(6):436. doi: 10.3390/jof7060436. J Fungi (Basel). 2021. PMID: 34070936 Free PMC article. Review.
-
Identification of long non-coding RNAs and RNA binding proteins in breast cancer subtypes.Sci Rep. 2022 Jan 13;12(1):693. doi: 10.1038/s41598-021-04664-z. Sci Rep. 2022. PMID: 35027621 Free PMC article.
References
-
- Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, et al. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci U S A. 2012;109(23):8983–8988. doi: 10.1073/pnas.1203244109. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

