Emerging Roles of DNA Glycosylases and the Base Excision Repair Pathway
- PMID: 31078398
- PMCID: PMC6699911
- DOI: 10.1016/j.tibs.2019.04.006
Emerging Roles of DNA Glycosylases and the Base Excision Repair Pathway
Abstract
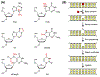
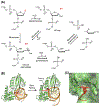
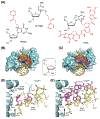
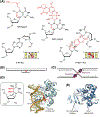
The base excision repair (BER) pathway historically has been associated with maintaining genome integrity by eliminating nucleobases with small chemical modifications. In the past several years, however, BER was found to play additional roles in genome maintenance and metabolism, including sequence-specific restriction modification and repair of bulky adducts and interstrand crosslinks. Central to this expanded biological utility are specialized DNA glycosylases - enzymes that selectively excise damaged, modified, or mismatched nucleobases. In this review we discuss the newly identified roles of the BER pathway and examine the structural and mechanistic features of the DNA glycosylases that enable these functions.
Keywords: DNA damage; DNA glycosylase; DNA repair; base excision repair; interstrand crosslink; secondary metabolite.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Structural Biology of the HEAT-Like Repeat Family of DNA Glycosylases.Bioessays. 2018 Nov;40(11):e1800133. doi: 10.1002/bies.201800133. Epub 2018 Sep 28. Bioessays. 2018. PMID: 30264543 Review.
-
NEIL3: A unique DNA glycosylase involved in interstrand DNA crosslink repair.DNA Repair (Amst). 2024 Jul;139:103680. doi: 10.1016/j.dnarep.2024.103680. Epub 2024 Apr 21. DNA Repair (Amst). 2024. PMID: 38663144 Free PMC article. Review.
-
The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions.Nature. 2015 Nov 12;527(7577):254-8. doi: 10.1038/nature15728. Epub 2015 Oct 28. Nature. 2015. PMID: 26524531 Free PMC article.
-
Initiating base excision repair in chromatin.DNA Repair (Amst). 2018 Nov;71:87-92. doi: 10.1016/j.dnarep.2018.08.011. Epub 2018 Aug 24. DNA Repair (Amst). 2018. PMID: 30170831 Free PMC article. Review.
-
Kinetic Methods for Studying DNA Glycosylases Functioning in Base Excision Repair.Methods Enzymol. 2017;592:357-376. doi: 10.1016/bs.mie.2017.03.016. Epub 2017 Apr 26. Methods Enzymol. 2017. PMID: 28668127 Free PMC article.
Cited by
-
Key Amino Acid Residues of Mitochondrial Transcription Factor A Synergize with Abasic (AP) Site Dynamics To Facilitate AP-Lyase Reactions.ACS Chem Biol. 2023 May 19;18(5):1168-1179. doi: 10.1021/acschembio.3c00047. Epub 2023 Mar 17. ACS Chem Biol. 2023. PMID: 36930463 Free PMC article.
-
Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1.DNA Repair (Amst). 2020 Jan;85:102741. doi: 10.1016/j.dnarep.2019.102741. Epub 2019 Nov 2. DNA Repair (Amst). 2020. PMID: 31733589 Free PMC article.
-
The structure of the Thermococcus gammatolerans McrB N-terminal domain reveals a new mode of substrate recognition and specificity among McrB homologs.J Biol Chem. 2020 Jan 17;295(3):743-756. doi: 10.1074/jbc.RA119.010188. Epub 2019 Dec 10. J Biol Chem. 2020. PMID: 31822563 Free PMC article.
-
Structural and Evolutionary Adaptations of Nei-Like DNA Glycosylases Proteins Involved in Base Excision Repair of Oxidative DNA Damage in Vertebrates.Oxid Med Cell Longev. 2022 Apr 4;2022:1144387. doi: 10.1155/2022/1144387. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35419164 Free PMC article.
-
Mycobacterial helicase Lhr abets resistance to DNA crosslinking agents mitomycin C and cisplatin.Nucleic Acids Res. 2023 Jan 11;51(1):218-235. doi: 10.1093/nar/gkac1222. Nucleic Acids Res. 2023. PMID: 36610794 Free PMC article.
References
-
- Friedberg EC et al. (2006) DNA Repair and Mutagenesis, 2nd edn., ASM Press.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

