Where is the right path heading from the centromere to spindle microtubules?
- PMID: 31075048
- PMCID: PMC6592241
- DOI: 10.1080/15384101.2019.1617008
Where is the right path heading from the centromere to spindle microtubules?
Abstract
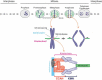
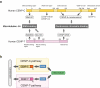
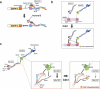
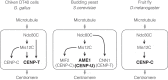
The kinetochore is a large protein complex that ensures accurate chromosome segregation during mitosis by connecting the centromere and spindle microtubules. One of the kinetochore sub-complexes, the constitutive centromere-associated network (CCAN), associates with the centromere and recruits another sub-complex, the KMN (KNL1, Mis12, and Ndc80 complexes) network (KMN), which binds to spindle microtubules. The CCAN-KMN interaction is mediated by two parallel pathways (CENP-C- and CENP-T-pathways) in the kinetochore, which bridge the centromere and microtubules. Here, we discuss dynamic protein-interaction changes in the two pathways that couple the centromere with spindle microtubules during mitotic progression.
Keywords: CCAN; KMN; Kinetochore; mitosis.
Figures

Similar articles
-
Multiple phosphorylations control recruitment of the KMN network onto kinetochores.Nat Cell Biol. 2018 Dec;20(12):1378-1388. doi: 10.1038/s41556-018-0230-0. Epub 2018 Nov 12. Nat Cell Biol. 2018. PMID: 30420662
-
Centromere Dysfunction Compromises Mitotic Spindle Pole Integrity.Curr Biol. 2019 Sep 23;29(18):3072-3080.e5. doi: 10.1016/j.cub.2019.07.052. Epub 2019 Sep 5. Curr Biol. 2019. PMID: 31495582
-
Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80.Mol Cell. 2018 Sep 20;71(6):923-939.e10. doi: 10.1016/j.molcel.2018.07.038. Epub 2018 Aug 30. Mol Cell. 2018. PMID: 30174292 Free PMC article.
-
Molecular architecture of vertebrate kinetochores.Exp Cell Res. 2012 Jul 15;318(12):1367-74. doi: 10.1016/j.yexcr.2012.02.016. Epub 2012 Feb 25. Exp Cell Res. 2012. PMID: 22391098 Review.
-
The kinetochore-microtubule interface at a glance.J Cell Sci. 2018 Aug 16;131(16):jcs214577. doi: 10.1242/jcs.214577. J Cell Sci. 2018. PMID: 30115751 Free PMC article. Review.
Cited by
-
Kinetochore Architecture Employs Diverse Linker Strategies Across Evolution.Front Cell Dev Biol. 2022 Jun 20;10:862637. doi: 10.3389/fcell.2022.862637. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35800888 Free PMC article. Review.
-
Recruitment of two Ndc80 complexes via the CENP-T pathway is sufficient for kinetochore functions.Nat Commun. 2022 Feb 14;13(1):851. doi: 10.1038/s41467-022-28403-8. Nat Commun. 2022. PMID: 35165266 Free PMC article.
-
The Role of Intravesicular Proteins and the Protein Corona of Extracellular Vesicles in the Development of Drug-Induced Polyneuropathy.Curr Issues Mol Biol. 2023 Apr 7;45(4):3302-3314. doi: 10.3390/cimb45040216. Curr Issues Mol Biol. 2023. PMID: 37185740 Free PMC article. Review.
-
Dynamics of kinetochore structure and its regulations during mitotic progression.Cell Mol Life Sci. 2020 Aug;77(15):2981-2995. doi: 10.1007/s00018-020-03472-4. Epub 2020 Feb 12. Cell Mol Life Sci. 2020. PMID: 32052088 Free PMC article. Review.
-
Vincristine-Induced Peripheral Neuropathy (VIPN) in Pediatric Tumors: Mechanisms, Risk Factors, Strategies of Prevention and Treatment.Int J Mol Sci. 2021 Apr 16;22(8):4112. doi: 10.3390/ijms22084112. Int J Mol Sci. 2021. PMID: 33923421 Free PMC article. Review.
References
-
- Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol. 2015. October 19;25(20):R1002–1018. - PubMed
-
- Sacristan C, Kops GJPL. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol. 2015. January;25(1):21–28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
