A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle
- PMID: 31071189
- PMCID: PMC6508626
- DOI: 10.1371/journal.ppat.1007736
A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle
Abstract
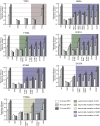
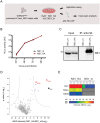
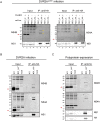
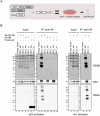
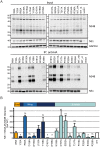
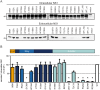
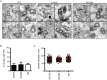
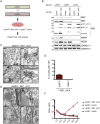
Dengue virus (DENV) has emerged as major human pathogen. Despite the serious socio-economic impact of DENV-associated diseases, antiviral therapy is missing. DENV replicates in the cytoplasm of infected cells and induces a membranous replication organelle, formed by invaginations of the endoplasmic reticulum membrane and designated vesicle packets (VPs). Nonstructural protein 1 (NS1) of DENV is a multifunctional protein. It is secreted from cells to counteract antiviral immune responses, but also critically contributes to the severe clinical manifestations of dengue. In addition, NS1 is indispensable for viral RNA replication, but the underlying molecular mechanism remains elusive. In this study, we employed a combination of genetic, biochemical and imaging approaches to dissect the determinants in NS1 contributing to its various functions in the viral replication cycle. Several important observations were made. First, we identified a cluster of amino acid residues in the exposed region of the β-ladder domain of NS1 that are essential for NS1 secretion. Second, we revealed a novel interaction of NS1 with the NS4A-2K-4B cleavage intermediate, but not with mature NS4A or NS4B. This interaction is required for RNA replication, with two residues within the connector region of the NS1 "Wing" domain being crucial for binding of the NS4A-2K-4B precursor. By using a polyprotein expression system allowing the formation of VPs in the absence of viral RNA replication, we show that the NS1 -NS4A-2K-4B interaction is not required for VP formation, arguing that the association between these two proteins plays a more direct role in the RNA amplification process. Third, through analysis of polyproteins containing deletions in NS1, and employing a trans-complementation assay, we show that both cis and trans acting elements within NS1 contribute to VP formation, with the capability of NS1 mutants to form VPs correlating with their capability to support RNA replication. In conclusion, these results reveal a direct role of NS1 in VP formation that is independent from RNA replication, and argue for a critical function of a previously unrecognized NS4A-2K-NS4B precursor specifically interacting with NS1 and promoting viral RNA replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Determinants in Nonstructural Protein 4A of Dengue Virus Required for RNA Replication and Replication Organelle Biogenesis.J Virol. 2021 Oct 13;95(21):e0131021. doi: 10.1128/JVI.01310-21. Epub 2021 Aug 11. J Virol. 2021. PMID: 34379504 Free PMC article.
-
Pan-serotype dengue virus inhibitor JNJ-A07 targets NS4A-2K-NS4B interaction with NS2B/NS3 and blocks replication organelle formation.Nat Commun. 2024 Jul 19;15(1):6080. doi: 10.1038/s41467-024-50437-3. Nat Commun. 2024. PMID: 39030239 Free PMC article.
-
Characterization of dengue virus NS4A and NS4B protein interaction.J Virol. 2015 Apr;89(7):3455-70. doi: 10.1128/JVI.03453-14. Epub 2015 Jan 7. J Virol. 2015. PMID: 25568208 Free PMC article.
-
Dengue virus NS2 and NS4: Minor proteins, mammoth roles.Biochem Pharmacol. 2018 Aug;154:54-63. doi: 10.1016/j.bcp.2018.04.008. Epub 2018 Apr 17. Biochem Pharmacol. 2018. PMID: 29674002 Review.
-
The dengue virus non-structural 1 protein: risks and benefits.Virus Res. 2014 Mar 6;181:53-60. doi: 10.1016/j.virusres.2014.01.001. Epub 2014 Jan 13. Virus Res. 2014. PMID: 24434336 Review.
Cited by
-
A Non-Replicative Role of the 3' Terminal Sequence of the Dengue Virus Genome in Membranous Replication Organelle Formation.Cell Rep. 2020 Jul 7;32(1):107859. doi: 10.1016/j.celrep.2020.107859. Cell Rep. 2020. PMID: 32640225 Free PMC article.
-
Zika NS1-induced ER remodeling is essential for viral replication.J Cell Biol. 2020 Feb 3;219(2):e201903062. doi: 10.1083/jcb.201903062. J Cell Biol. 2020. PMID: 31868887 Free PMC article.
-
Attenuation of Zika Virus by Passage in Human HeLa Cells.Vaccines (Basel). 2019 Aug 20;7(3):93. doi: 10.3390/vaccines7030093. Vaccines (Basel). 2019. PMID: 31434319 Free PMC article.
-
Pathogenesis and virulence of flavivirus infections.Virulence. 2021 Dec;12(1):2814-2838. doi: 10.1080/21505594.2021.1996059. Virulence. 2021. PMID: 34696709 Free PMC article. Review.
-
HEATR5B associates with dynein-dynactin and promotes motility of AP1-bound endosomal membranes.EMBO J. 2023 Dec 1;42(23):e114473. doi: 10.15252/embj.2023114473. Epub 2023 Oct 24. EMBO J. 2023. PMID: 37872872 Free PMC article.
References
-
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI (2013) The global distribution and burden of dengue. Nature 496: 504–507. 10.1038/nature12060 - DOI - PMC - PubMed
-
- World Health Organization (2016) Dengue vaccine: WHO position paper—July 2016. Wkly Epidemiol Rec 91: 349–364. - PubMed
-
- Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R (2009) Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5: 365–375. 10.1016/j.chom.2009.03.007 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

