Lymphoid-specific helicase promotes the growth and invasion of hepatocellular carcinoma by transcriptional regulation of centromere protein F expression
- PMID: 31066149
- PMCID: PMC6609811
- DOI: 10.1111/cas.14037
Lymphoid-specific helicase promotes the growth and invasion of hepatocellular carcinoma by transcriptional regulation of centromere protein F expression
Abstract
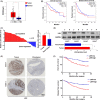
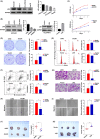
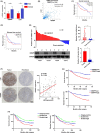
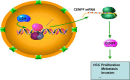
Lymphoid-specific helicase (LSH) is overexpressed in tumor tissues and its overexpression is associated with poor prognosis in several cancers. However, the role and molecular mechanism of LSH in hepatocellular carcinoma (HCC) remains largely unknown. Herein, we report that LSH was overexpressed in tumor tissues of HCC, and overexpression of LSH was associated with poor prognosis from a public HCC database, and validated by clinical samples from our department. Ectopic LSH expression promoted the growth of HCC cells in vivo and in vitro. Mechanistically, LSH overexpression promoted tumor growth by activating transcription of centromere protein F (CENPF). Clinically, overexpression of LSH and/or CENPF correlated with shorter overall survival and higher cumulative recurrence rates of HCC. In conclusion, LSH promotes tumor growth of HCC through transcriptional regulation of CENPF expression. Therefore, LSH may be a novel predictor for prognosis and a potential therapeutic target for HCC.
Keywords: CENPF; ChIP-seq; HCC; LSH; RNA-seq.
© 2019 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Figures


Similar articles
-
Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma.J Hepatol. 2018 Jun;68(6):1214-1227. doi: 10.1016/j.jhep.2018.01.012. Epub 2018 Jan 31. J Hepatol. 2018. PMID: 29378234
-
Characterization of the oncogenic function of centromere protein F in hepatocellular carcinoma.Biochem Biophys Res Commun. 2013 Jul 12;436(4):711-8. doi: 10.1016/j.bbrc.2013.06.021. Epub 2013 Jun 17. Biochem Biophys Res Commun. 2013. PMID: 23791740
-
Centromere protein F promotes progression of hepatocellular carcinoma through ERK and cell cycle-associated pathways.Cancer Gene Ther. 2022 Jul;29(7):1033-1042. doi: 10.1038/s41417-021-00404-7. Epub 2021 Dec 2. Cancer Gene Ther. 2022. PMID: 34857915
-
Actin-like 6A predicts poor prognosis of hepatocellular carcinoma and promotes metastasis and epithelial-mesenchymal transition.Hepatology. 2016 Apr;63(4):1256-71. doi: 10.1002/hep.28417. Epub 2016 Jan 26. Hepatology. 2016. PMID: 26698646 Free PMC article.
-
Lymphoid-specific helicase in epigenetics, DNA repair and cancer.Br J Cancer. 2022 Feb;126(2):165-173. doi: 10.1038/s41416-021-01543-2. Epub 2021 Sep 7. Br J Cancer. 2022. PMID: 34493821 Free PMC article. Review.
Cited by
-
Centromere Protein F (CENPF) Serves as a Potential Prognostic Biomarker and Target for Human Hepatocellular Carcinoma.J Cancer. 2021 Mar 15;12(10):2933-2951. doi: 10.7150/jca.52187. eCollection 2021. J Cancer. 2021. PMID: 33854594 Free PMC article.
-
CENPF/CDK1 signaling pathway enhances the progression of adrenocortical carcinoma by regulating the G2/M-phase cell cycle.J Transl Med. 2022 Feb 5;20(1):78. doi: 10.1186/s12967-022-03277-y. J Transl Med. 2022. PMID: 35123514 Free PMC article.
-
Identification of a Prognostic Model Based on 2-Gene Signature and Analysis of Corresponding Tumor Microenvironment in Alcohol-Related Hepatocellular Carcinoma.Front Oncol. 2021 Sep 27;11:719355. doi: 10.3389/fonc.2021.719355. eCollection 2021. Front Oncol. 2021. PMID: 34646769 Free PMC article.
-
Role of CENPF and NDC80 in the rehabilitation nursing of hepatocellular carcinoma and cirrhosis: An observational study.Medicine (Baltimore). 2024 May 3;103(18):e37984. doi: 10.1097/MD.0000000000037984. Medicine (Baltimore). 2024. PMID: 38701255 Free PMC article.
-
Integrative analysis identifies key mRNA biomarkers for diagnosis, prognosis, and therapeutic targets of HCV-associated hepatocellular carcinoma.Aging (Albany NY). 2021 May 4;13(9):12865-12895. doi: 10.18632/aging.202957. Epub 2021 May 4. Aging (Albany NY). 2021. PMID: 33946043 Free PMC article.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. - PubMed
MeSH terms
Substances
Grants and funding
- 17411951200/Shanghai Municipal Natural Science Foundation
- 18410720700/Shanghai Municipal Natural Science Foundation
- PWYgy2018-02/the Outstanding Clinical Discipline Project of Shanghai Pudong
- CBSKL201717/Cancer Biology State Key Laboratory Project
- 81472840/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical

