An Overview of Methods for Reconstructing 3-D Chromosome and Genome Structures from Hi-C Data
- PMID: 31049033
- PMCID: PMC6482566
- DOI: 10.1186/s12575-019-0094-0
An Overview of Methods for Reconstructing 3-D Chromosome and Genome Structures from Hi-C Data
Abstract
Over the past decade, methods for predicting three-dimensional (3-D) chromosome and genome structures have proliferated. This has been primarily due to the development of high-throughput, next-generation chromosome conformation capture (3C) technologies, which have provided next-generation sequencing data about chromosome conformations in order to map the 3-D genome structure. The introduction of the Hi-C technique-a variant of the 3C method-has allowed researchers to extract the interaction frequency (IF) for all loci of a genome at high-throughput and at a genome-wide scale. In this review we describe, categorize, and compare the various methods developed to map chromosome and genome structures from 3C data-particularly Hi-C data. We summarize the improvements introduced by these methods, describe the approach used for method evaluation, and discuss how these advancements shape the future of genome structure construction.
Keywords: 3-D chromosome and genome structure; 3-D genome; Chromosome conformation capture; Contact-based modeling; Distance-based modeling; Hi-C; Optimization.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
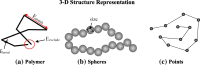
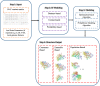
Similar articles
-
Tethered Chromosome Conformation Capture Sequencing in Triticeae: A Valuable Tool for Genome Assembly.Bio Protoc. 2018 Aug 5;8(15):e2955. doi: 10.21769/BioProtoc.2955. eCollection 2018 Aug 5. Bio Protoc. 2018. PMID: 34395764 Free PMC article.
-
A maximum likelihood algorithm for reconstructing 3D structures of human chromosomes from chromosomal contact data.BMC Genomics. 2018 Feb 23;19(1):161. doi: 10.1186/s12864-018-4546-8. BMC Genomics. 2018. PMID: 29471801 Free PMC article.
-
Hi-C 3.0: Improved Protocol for Genome-Wide Chromosome Conformation Capture.Curr Protoc. 2021 Jul;1(7):e198. doi: 10.1002/cpz1.198. Curr Protoc. 2021. PMID: 34286910 Free PMC article.
-
3C and 3C-based techniques: the powerful tools for spatial genome organization deciphering.Mol Cytogenet. 2018 Mar 9;11:21. doi: 10.1186/s13039-018-0368-2. eCollection 2018. Mol Cytogenet. 2018. PMID: 29541161 Free PMC article. Review.
-
Chromosome conformation capture technologies and their impact in understanding genome function.Chromosoma. 2017 Feb;126(1):33-44. doi: 10.1007/s00412-016-0593-6. Epub 2016 Apr 30. Chromosoma. 2017. PMID: 27130552 Review.
Cited by
-
Superstructure Detection in Nucleosome Distribution Shows Common Pattern within a Chromosome and within the Genome.Life (Basel). 2022 Apr 6;12(4):541. doi: 10.3390/life12040541. Life (Basel). 2022. PMID: 35455033 Free PMC article.
-
Reconstruct high-resolution 3D genome structures for diverse cell-types using FLAMINGO.Nat Commun. 2022 May 12;13(1):2645. doi: 10.1038/s41467-022-30270-2. Nat Commun. 2022. PMID: 35551182 Free PMC article.
-
Nuclear genome organization in fungi: from gene folding to Rabl chromosomes.FEMS Microbiol Rev. 2023 May 19;47(3):fuad021. doi: 10.1093/femsre/fuad021. FEMS Microbiol Rev. 2023. PMID: 37197899 Free PMC article.
-
Depletion of lamins B1 and B2 promotes chromatin mobility and induces differential gene expression by a mesoscale-motion-dependent mechanism.Genome Biol. 2024 Mar 22;25(1):77. doi: 10.1186/s13059-024-03212-y. Genome Biol. 2024. PMID: 38519987 Free PMC article.
-
GSDB: a database of 3D chromosome and genome structures reconstructed from Hi-C data.BMC Mol Cell Biol. 2020 Aug 5;21(1):60. doi: 10.1186/s12860-020-00304-y. BMC Mol Cell Biol. 2020. PMID: 32758136 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

