A novel role of U1 snRNP: Splice site selection from a distance
- PMID: 31042550
- PMCID: PMC6557577
- DOI: 10.1016/j.bbagrm.2019.04.004
A novel role of U1 snRNP: Splice site selection from a distance
Abstract
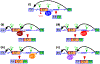
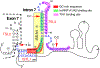
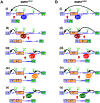
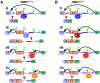
Removal of introns by pre-mRNA splicing is fundamental to gene function in eukaryotes. However, understanding the mechanism by which exon-intron boundaries are defined remains a challenging endeavor. Published reports support that the recruitment of U1 snRNP at the 5'ss marked by GU dinucleotides defines the 5'ss as well as facilitates 3'ss recognition through cross-exon interactions. However, exceptions to this rule exist as U1 snRNP recruited away from the 5'ss retains the capability to define the splice site, where the cleavage takes place. Independent reports employing exon 7 of Survival Motor Neuron (SMN) genes suggest a long-distance effect of U1 snRNP on splice site selection upon U1 snRNP recruitment at target sequences with or without GU dinucleotides. These findings underscore that sequences distinct from the 5'ss may also impact exon definition if U1 snRNP is recruited to them through partial complementarity with the U1 snRNA. In this review we discuss the expanded role of U1 snRNP in splice-site selection due to U1 ability to be recruited at more sites than predicted solely based on GU dinucleotides.
Keywords: Cryptic splice site; ISS-N1; SMA; SMN; Splicing; U1 snRNP.
Copyright © 2019 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest Statement:
Authors declare no conflict of interest.
Figures
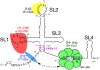
Similar articles
-
Principles and correction of 5'-splice site selection.RNA Biol. 2022 Jan;19(1):943-960. doi: 10.1080/15476286.2022.2100971. RNA Biol. 2022. PMID: 35866748 Free PMC article. Review.
-
Activation of a cryptic 5' splice site reverses the impact of pathogenic splice site mutations in the spinal muscular atrophy gene.Nucleic Acids Res. 2017 Dec 1;45(21):12214-12240. doi: 10.1093/nar/gkx824. Nucleic Acids Res. 2017. PMID: 28981879 Free PMC article.
-
Profiling of cis- and trans-acting factors supporting noncanonical splice site activation.RNA Biol. 2021 Jan;18(1):118-130. doi: 10.1080/15476286.2020.1798111. Epub 2020 Aug 5. RNA Biol. 2021. PMID: 32693676 Free PMC article.
-
U1 snRNA promotes the selection of nearby 5' splice sites by U6 snRNA in mammalian cells.Genes Dev. 1996 Feb 1;10(3):338-50. doi: 10.1101/gad.10.3.338. Genes Dev. 1996. PMID: 8595884
-
Recognition of the 5' splice site by the spliceosome.Acta Biochim Pol. 1998;45(4):869-81. Acta Biochim Pol. 1998. PMID: 10397335 Review.
Cited by
-
VRK1 (Y213H) homozygous mutant impairs Cajal bodies in a hereditary case of distal motor neuropathy.Ann Clin Transl Neurol. 2020 May;7(5):808-818. doi: 10.1002/acn3.51050. Epub 2020 May 4. Ann Clin Transl Neurol. 2020. PMID: 32365420 Free PMC article.
-
Noncoding RNAs regulate alternative splicing in Cancer.J Exp Clin Cancer Res. 2021 Jan 6;40(1):11. doi: 10.1186/s13046-020-01798-2. J Exp Clin Cancer Res. 2021. PMID: 33407694 Free PMC article. Review.
-
Mechanisms of Neuronal Alternative Splicing and Strategies for Therapeutic Interventions.J Neurosci. 2019 Oct 16;39(42):8193-8199. doi: 10.1523/JNEUROSCI.1149-19.2019. J Neurosci. 2019. PMID: 31619487 Free PMC article. Review.
-
HIV-1: To Splice or Not to Splice, That Is the Question.Viruses. 2021 Jan 26;13(2):181. doi: 10.3390/v13020181. Viruses. 2021. PMID: 33530363 Free PMC article. Review.
-
How RNA structure dictates the usage of a critical exon of spinal muscular atrophy gene.Biochim Biophys Acta Gene Regul Mech. 2019 Nov-Dec;1862(11-12):194403. doi: 10.1016/j.bbagrm.2019.07.004. Epub 2019 Jul 16. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31323435 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

