Repurposing Ponatinib as a Potent Agent against KIT Mutant Melanomas
- PMID: 31037149
- PMCID: PMC6485277
- DOI: 10.7150/thno.30890
Repurposing Ponatinib as a Potent Agent against KIT Mutant Melanomas
Abstract
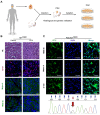
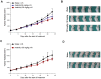
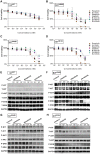
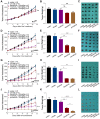
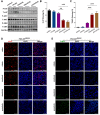
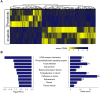
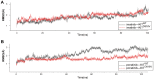
Rationale: Mutations in KIT, a major cancer driver gene, are now considered as important drug targets for the treatment of melanomas arising from mucosal and acral tissues and from chronically sun-damaged sites. At present, imatinib is the only targeted drug for KIT-mutation-bearing melanomas that is recommended by the National Comprehensive Cancer Network (NCCN) Clinical Practice guidelines. Patients with KIT mutations, however, are either insensitive or rapidly progress to imatinib insensitivity, which restricts its clinical use. Thus, effective inhibitors of KIT-mutation-bearing melanomas are urgently needed. Methods: A cohort of patient-derived tumor xenograft (PDX) models and corresponding PDX-derived cells (PDCs) from patients with melanomas harboring KIT mutations (KITV560D, KITK642E and KITD816V) were established, characterized, and then used to test the in vitro and, subsequently, in vivo inhibitory effects of a panel of known KIT inhibitors. Results: Ponatinib was more potent than imatinib against cells bearing KIT mutations. In vivo drug efficacy evaluation experiments showed that ponatinib treatment caused much stronger inhibition of KIT-mutation-bearing melanomas than did imatinib. Mechanistically, molecular dynamics (MD) simulations revealed a plausible atomic-level explanation for the observation that ponatinib has a higher affinity for the KITD816V mutant protein than does imatinib. Conclusions: Our study of KIT-mutation-and KITWT-bearing melanomas demonstrates that ponatinib is a far more potent inhibitor than is imatinib for KIT-mutation-bearing melanomas and thus underscores that ponatinib should be given priority consideration for the design of precision treatments for melanoma patients triaged to have KIT mutations. Moreover, our work provides a rationale for undertaking clinical trials to examine the repurposing of ponatinib, which is already approved for use in leukemia, for use in treating a large subset of melanoma patients.
Keywords: KIT; melanomas; patient-derived xenograft models; ponatinib.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
Similar articles
-
Ponatinib induces apoptosis in imatinib-resistant human mast cells by dephosphorylating mutant D816V KIT and silencing β-catenin signaling.Mol Cancer Ther. 2014 May;13(5):1217-30. doi: 10.1158/1535-7163.MCT-13-0397. Epub 2014 Feb 19. Mol Cancer Ther. 2014. PMID: 24552773
-
Ponatinib inhibits polyclonal drug-resistant KIT oncoproteins and shows therapeutic potential in heavily pretreated gastrointestinal stromal tumor (GIST) patients.Clin Cancer Res. 2014 Nov 15;20(22):5745-5755. doi: 10.1158/1078-0432.CCR-14-1397. Epub 2014 Sep 19. Clin Cancer Res. 2014. PMID: 25239608 Free PMC article.
-
Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin.J Clin Oncol. 2013 Sep 10;31(26):3182-90. doi: 10.1200/JCO.2012.47.7836. Epub 2013 Jun 17. J Clin Oncol. 2013. PMID: 23775962 Free PMC article. Clinical Trial.
-
Therapeutic implications of KIT in melanoma.Cancer J. 2012 Mar-Apr;18(2):137-41. doi: 10.1097/PPO.0b013e31824b2404. Cancer J. 2012. PMID: 22453014 Review.
-
The role of small molecule Kit protein-tyrosine kinase inhibitors in the treatment of neoplastic disorders.Pharmacol Res. 2018 Jul;133:35-52. doi: 10.1016/j.phrs.2018.04.020. Epub 2018 Apr 25. Pharmacol Res. 2018. PMID: 29704617 Review.
Cited by
-
Resistance to Molecularly Targeted Therapies in Melanoma.Cancers (Basel). 2021 Mar 5;13(5):1115. doi: 10.3390/cancers13051115. Cancers (Basel). 2021. PMID: 33807778 Free PMC article. Review.
-
Novel biomolecules in targeted cancer therapy: a new approach towards precision medicine.Med Oncol. 2023 Oct 7;40(11):323. doi: 10.1007/s12032-023-02168-6. Med Oncol. 2023. PMID: 37804361 Review.
-
The MNK1/2-eIF4E Axis as a Potential Therapeutic Target in Melanoma.Int J Mol Sci. 2020 Jun 5;21(11):4055. doi: 10.3390/ijms21114055. Int J Mol Sci. 2020. PMID: 32517051 Free PMC article. Review.
-
The efficacy and safety of dalpiciclib, a cyclin-dependent kinase 4/6 inhibitor, in patients with advanced head and neck mucosal melanoma harboring CDK4 amplification.BMC Med. 2024 May 29;22(1):215. doi: 10.1186/s12916-024-03431-x. BMC Med. 2024. PMID: 38807144 Free PMC article.
-
Melanoma Targeted Therapies beyond BRAF-Mutant Melanoma: Potential Druggable Mutations and Novel Treatment Approaches.Cancers (Basel). 2021 Nov 22;13(22):5847. doi: 10.3390/cancers13225847. Cancers (Basel). 2021. PMID: 34831002 Free PMC article. Review.
References
-
- Ossio R, Roldán-Marín R, Martínez-Said H, Adams DJ, Robles-Espinoza CD. Melanoma: a global perspective. Nat Rev Cancer. 2017;17:393. - PubMed
-
- Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–6. - PubMed
-
- Todd JR, Scurr LL, Becker TM, Kefford RF, Rizos H. The MAPK pathway functions as a redundant survival signal that reinforces the PI3K cascade in c-Kit mutant melanoma. Oncogene. 2014;33:236–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

