Three New Cs for CRISPR: Collateral, Communicate, Cooperate
- PMID: 31036344
- PMCID: PMC6525018
- DOI: 10.1016/j.tig.2019.03.009
Three New Cs for CRISPR: Collateral, Communicate, Cooperate
Abstract
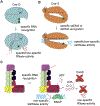
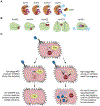
Clustered regularly interspaced short palindromic repeats (CRISPR) loci and their associated (cas) genes provide protection against invading phages and plasmids in prokaryotes. Typically, short sequences are captured from the genome of the invader, integrated into the CRISPR locus, and transcribed into short RNAs that direct RNA-guided Cas nucleases to the nucleic acids of the invader for their degradation. Recent work in the field has revealed unexpected features of the CRISPR-Cas mechanism: (i) collateral, nonspecific, cleavage of host nucleic acids; (ii) secondary messengers that amplify the immune response; and (iii) immunosuppression of CRISPR targeting by phage-encoded inhibitors. Here, we review these new and exciting findings.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures
Similar articles
-
RNA Guide Complementarity Prevents Self-Targeting in Type VI CRISPR Systems.Mol Cell. 2018 Sep 6;71(5):791-801.e3. doi: 10.1016/j.molcel.2018.07.013. Epub 2018 Aug 16. Mol Cell. 2018. PMID: 30122537 Free PMC article.
-
Cooperation between Different CRISPR-Cas Types Enables Adaptation in an RNA-Targeting System.mBio. 2021 Mar 30;12(2):e03338-20. doi: 10.1128/mBio.03338-20. mBio. 2021. PMID: 33785624 Free PMC article.
-
Unity among the diverse RNA-guided CRISPR-Cas interference mechanisms.J Biol Chem. 2024 Jun;300(6):107295. doi: 10.1016/j.jbc.2024.107295. Epub 2024 Apr 18. J Biol Chem. 2024. PMID: 38641067 Free PMC article. Review.
-
High-Throughput Characterization of Cascade type I-E CRISPR Guide Efficacy Reveals Unexpected PAM Diversity and Target Sequence Preferences.Genetics. 2017 Aug;206(4):1727-1738. doi: 10.1534/genetics.117.202580. Epub 2017 Jun 20. Genetics. 2017. PMID: 28634160 Free PMC article.
-
Shooting the messenger: RNA-targetting CRISPR-Cas systems.Biosci Rep. 2018 Jun 21;38(3):BSR20170788. doi: 10.1042/BSR20170788. Print 2018 Jun 29. Biosci Rep. 2018. PMID: 29748239 Free PMC article. Review.
Cited by
-
Kinetic analysis of Cas12a and Cas13a RNA-Guided nucleases for development of improved CRISPR-Based diagnostics.iScience. 2021 Aug 18;24(9):102996. doi: 10.1016/j.isci.2021.102996. eCollection 2021 Sep 24. iScience. 2021. PMID: 34505008 Free PMC article.
-
The Cyclic Oligoadenylate Signaling Pathway of Type III CRISPR-Cas Systems.Front Microbiol. 2021 Jan 20;11:602789. doi: 10.3389/fmicb.2020.602789. eCollection 2020. Front Microbiol. 2021. PMID: 33552016 Free PMC article. Review.
-
Allosteric activation of CRISPR-Cas12a requires the concerted movement of the bridge helix and helix 1 of the RuvC II domain.Nucleic Acids Res. 2022 Sep 23;50(17):10153-10168. doi: 10.1093/nar/gkac767. Nucleic Acids Res. 2022. PMID: 36107767 Free PMC article.
-
Gene regulatory and gene editing tools and their applications for retinal diseases and neuroprotection: From proof-of-concept to clinical trial.Front Neurosci. 2022 Oct 20;16:924917. doi: 10.3389/fnins.2022.924917. eCollection 2022. Front Neurosci. 2022. PMID: 36340792 Free PMC article. Review.
-
Determinants of CRISPR Cas12a nuclease activation by DNA and RNA targets.Nucleic Acids Res. 2024 May 8;52(8):4502-4522. doi: 10.1093/nar/gkae152. Nucleic Acids Res. 2024. PMID: 38477377 Free PMC article.
References
-
- Barrangou R et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 (5819), 1709–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

