Complementary Activity of ETV5, RBPJ, and TCF3 Drives Formative Transition from Naive Pluripotency
- PMID: 31031137
- PMCID: PMC6509416
- DOI: 10.1016/j.stem.2019.03.017
Complementary Activity of ETV5, RBPJ, and TCF3 Drives Formative Transition from Naive Pluripotency
Abstract
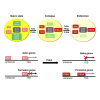
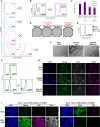
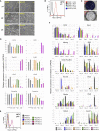
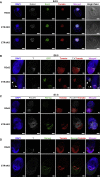
The gene regulatory network (GRN) of naive mouse embryonic stem cells (ESCs) must be reconfigured to enable lineage commitment. TCF3 sanctions rewiring by suppressing components of the ESC transcription factor circuitry. However, TCF3 depletion only delays and does not prevent transition to formative pluripotency. Here, we delineate additional contributions of the ETS-family transcription factor ETV5 and the repressor RBPJ. In response to ERK signaling, ETV5 switches activity from supporting self-renewal and undergoes genome relocation linked to commissioning of enhancers activated in formative epiblast. Independent upregulation of RBPJ prevents re-expression of potent naive factors, TBX3 and NANOG, to secure exit from the naive state. Triple deletion of Etv5, Rbpj, and Tcf3 disables ESCs, such that they remain largely undifferentiated and locked in self-renewal, even in the presence of differentiation stimuli. Thus, genetic elimination of three complementary drivers of network transition stalls developmental progression, emulating environmental insulation by small-molecule inhibitors.
Keywords: ETS factors; RBPJ; commitment; differentiation; embryonic stem cell; epiblast; gene regulatory network; pluripotency; self-renewal.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



Comment in
-
Pluripotency on Lockdown after Deletion of Three Transcription Regulators.Cell Stem Cell. 2019 May 2;24(5):681-683. doi: 10.1016/j.stem.2019.04.008. Cell Stem Cell. 2019. PMID: 31051130
Similar articles
-
The transcription factor OCT6 promotes the dissolution of the naïve pluripotent state by repressing Nanog and activating a formative state gene regulatory network.Sci Rep. 2024 May 7;14(1):10420. doi: 10.1038/s41598-024-59247-5. Sci Rep. 2024. PMID: 38710730 Free PMC article.
-
ERK signalling eliminates Nanog and maintains Oct4 to drive the formative pluripotency transition.Development. 2024 Jul 15;151(14):dev203106. doi: 10.1242/dev.203106. Epub 2024 Jul 26. Development. 2024. PMID: 39069943
-
Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity.Nat Cell Biol. 2018 May;20(5):565-574. doi: 10.1038/s41556-018-0086-3. Epub 2018 Apr 16. Nat Cell Biol. 2018. PMID: 29662175 Free PMC article.
-
Mapping the route from naive pluripotency to lineage specification.Philos Trans R Soc Lond B Biol Sci. 2014 Dec 5;369(1657):20130540. doi: 10.1098/rstb.2013.0540. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 25349449 Free PMC article. Review.
-
The transcriptional network controlling pluripotency in ES cells.Cold Spring Harb Symp Quant Biol. 2008;73:195-202. doi: 10.1101/sqb.2008.72.001. Cold Spring Harb Symp Quant Biol. 2008. PMID: 19478325 Review.
Cited by
-
The Nucleosome Remodelling and Deacetylation complex coordinates the transcriptional response to lineage commitment in pluripotent cells.Biol Open. 2024 Jan 15;13(1):bio060101. doi: 10.1242/bio.060101. Epub 2024 Jan 22. Biol Open. 2024. PMID: 38149716 Free PMC article.
-
Cnot8 eliminates naïve regulation networks and is essential for naïve-to-formative pluripotency transition.Nucleic Acids Res. 2022 May 6;50(8):4414-4435. doi: 10.1093/nar/gkac236. Nucleic Acids Res. 2022. PMID: 35390160 Free PMC article.
-
Integrative and perturbation-based analysis of the transcriptional dynamics of TGFβ/BMP system components in transition from embryonic stem cells to neural progenitors.Stem Cells. 2020 Feb;38(2):202-217. doi: 10.1002/stem.3111. Epub 2019 Dec 13. Stem Cells. 2020. PMID: 31675135 Free PMC article.
-
Epigenetics as "conductor" in "orchestra" of pluripotent states.Cell Tissue Res. 2022 Nov;390(2):141-172. doi: 10.1007/s00441-022-03667-0. Epub 2022 Jul 15. Cell Tissue Res. 2022. PMID: 35838826 Review.
-
β-Catenin safeguards the ground state of mousepluripotency by strengthening the robustness of the transcriptional apparatus.Sci Adv. 2020 Jul 17;6(29):eaba1593. doi: 10.1126/sciadv.aba1593. eCollection 2020 Jul. Sci Adv. 2020. PMID: 32832621 Free PMC article.
References
-
- Arber S., Ladle D.R., Lin J.H., Frank E., Jessell T.M. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

