Triggering MSR1 promotes JNK-mediated inflammation in IL-4-activated macrophages
- PMID: 31028084
- PMCID: PMC6545745
- DOI: 10.15252/embj.2018100299
Triggering MSR1 promotes JNK-mediated inflammation in IL-4-activated macrophages
Abstract
Alternatively activated M2 macrophages play an important role in maintenance of tissue homeostasis by scavenging dead cells, cell debris and lipoprotein aggregates via phagocytosis. Using proteomics, we investigated how alternative activation, driven by IL-4, modulated the phagosomal proteome to control macrophage function. Our data indicate that alternative activation enhances homeostatic functions such as proteolysis, lipolysis and nutrient transport. Intriguingly, we identified the enhanced recruitment of the TAK1/MKK7/JNK signalling complex to phagosomes of IL-4-activated macrophages. The recruitment of this signalling complex was mediated through K63 polyubiquitylation of the macrophage scavenger receptor 1 (MSR1). Triggering of MSR1 in IL-4-activated macrophages leads to enhanced JNK activation, thereby promoting a phenotypic switch from an anti-inflammatory to a pro-inflammatory state, which was abolished upon MSR1 deletion or JNK inhibition. Moreover, MSR1 K63 polyubiquitylation correlated with the activation of JNK signalling in ovarian cancer tissue from human patients, suggesting that it may be relevant for macrophage phenotypic shift in vivo Altogether, we identified that MSR1 signals through JNK via K63 polyubiquitylation and provides evidence for the receptor's involvement in macrophage polarization.
Keywords: macrophage scavenger receptor 1; phagosome; proteomics; scavenger receptor; tumour‐associated macrophages.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
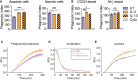
- A
Phagocytosis assay of apoptotic or necrotic GFP‐expressing RAW264.7 cells in M2 (IL‐4 or IL13) and untreated M0 BMDMs.
- B
Phagocytosis of fluorescent negatively charged carboxylated and positively charged amino microspheres (B) in primary M2 (IL‐4) and M0 macrophages. Cytochalasin D (Cyto) (6 μM) was used as an inhibitor of phagocytosis, 1 h before phagocytosis.
- C–E
Real‐time fluorescence assays for intraphagosomal proteolysis (C), acidification (D) and lipolysis (E) show substantially increased proteolysis, acidification and lipolysis in the phagosomes of M2 (IL‐4) macrophages. The kinetics of proteolysis, acidification and lipolysis of phagocytosed beads were plotted as a ratio of substrate fluorescence to calibration fluorescence. Beads were added to macrophages at 0 min. (E) is a representative of three independent experiments. Leupeptin (100 nM) and bafilomycin (100 nM) treatments serve as negative controls in (C) and (D), respectively.

Workflow of the phagosome proteomic experiment.
Volcano plot of the phagosome proteome data. 1,766 proteins were quantified of which 121 proteins were significantly up‐regulated and 62 proteins were down‐regulated in M2 (IL‐4) macrophages. Selected proteins are indicated.
Heatmap of proteomic data shows high reproducibility between biological replicates. Selected proteins are highlighted.
Selected Gene Ontology (GO) terms of biological processes significantly up‐regulated and down‐regulated on phagosomes of M2 (IL‐4) macrophages. Selected proteins of these GO‐terms are highlighted. Error bars represent standard deviations from three biological replicates.
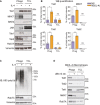
Immunoblot (IB) analysis showing the recruitment of TAK1, MKK7, JNK and TAB1/TAB2 to the phagosome in M2 (IL‐4) macrophages, while MKK4 is not recruited. Rab7a, a phagosomal marker, was used as a loading control for phagosomes, and vimentin was used as a loading control for total cell lysates. The white line in the JNK blot shows other bands were cut out. Full blot can be seen in Appendix Figure S4.
Quantitation of three independent IB experiments by ImageJ of non‐saturated blots for TAK1, MKK7, TAB1 and TAB2 expression on phagosomes of resting M0 macrophages and M2 (IL‐4) macrophages. Error bars represent SEM. *P < 0.01, **P < 0.001 (Student's t‐test). Three replicates were used.
IB showing enrichment of K63‐polyubiquitylated proteins on the phagosome of M2 (IL‐4) macrophages compared to M0 macrophages.
Treatment with the UBC13 inhibitor NSC697923 reduces recruitment of TAB1, TAB2, TAK1 and MKK7 to the phagosome of M2 (IL‐4) macrophages, indicating a K63‐polyubiquitylation‐dependent translocation for these proteins.

Workflow for TUBE pull‐down of K63‐polyubiquitylated proteins from phagosomal extracts.
Selected ubiquitylated phagosomal proteins identified by TUBE‐MS approach.
Sequences alignment of N‐terminal region of murine and human MSR1 shows high‐sequence identity and conserved ubiquitylated lysine. The arrow points out the ubiquitylated lysine residue.
Tab2‐TUBE pull‐downs from phagosome extracts of M2 (IL‐4) macrophages treated with the K63‐specific deubiquitylase (DUB) AMSH‐LP or the unspecific DUB USP2.
MSR1 immunoprecipitation from M2 (IL‐4) macrophage phagosomes shows that TAB1, TAB2, TAK1 and MKK7 bind to polyubiquitylated MSR1. TAK1 shows a specific pattern of post‐translational modifications indicative of its activation.

Immunoblot against MSR1 of TUBE pull‐downs of MSR1 WT and KO BMDMs shows increasing amounts of ubiquitylated MSR1 upon alternative activation and further increases upon MSR1 ligation with fucoidan or oxLDL.
Msr1 +/+ (WT) and Msr1 −/− (KO) M0 macrophages and M2 (IL‐4) macrophages untreated or stimulated with the MSR1 ligands fucoidan or oxLDL (50 μg/ml, 30 min) were analysed for the phosphorylated and the total forms of JNK1/2 and MSR1. ITGAM (CD11b) serves as a loading control. Both fucoidan and oxLDL activate JNK in a MSR1‐dependent manner.
IL4‐activated MSR1 knock‐out BMDMs were transfected with WT or K27R MSR1 and treated with 50 μg/ml fucoidan for 1 h. Mutation of the ubiquitylation site K27 abolishes MSR1 signalling.
qPCR data of Tnfa, Il1b and Ccl2 mRNA levels in WT and MSR1 KO M0 and M2 (IL‐4) BMDMs show an MSR1‐dependent increase in pro‐inflammatory cytokines in response to MSR1 ligation by fucoidan.
Inhibition of JNK by JNK‐IN8 reduces expression of Tnfa and Ccl2 upon MSR1 ligation, showing that it is JNK‐dependent.
Flow cytometry analysis of cell surface markers in WT and MSR1 −/− M0 macrophages and M2 (IL‐4) macrophages untreated or stimulated with the MSR1 ligands fucoidan (50 μg/ml, 24 h). Data show MSR1‐dependent increase in the early activation markers CD54, CD69 and CD86 and a decrease in the M2 marker CD301b/Mgl2. CD11b serves as a control.

Immunohistochemistry analysis of MSR1 in patient ovarian, uterus and sarcoma cancers showing tumour‐associated macrophages.
Pull‐downs of K63‐polyubiquitin chains and IB analysis of five human primary cancers show a correlation between the amount of polyubiquitylated MSR1 and JNK activation in an ovarian tumour.
Working model: MSR1 is activated by ligation through many different substrates, including apoptotic cells, fucoidan or oxidized LDL (Signal 1). However, only when the macrophage is IL‐4‐activated (Signal 2) becomes MSR1 ubiquitylated by an unknown E2/E3 ligase. This ubiquitylation recruits Tab2/3, Tak1, Mkk7 and finally JNK, thereby allowing MSR1 to signal directly through the JNK signalling pathway which induces pro‐inflammatory gene transcription.
Similar articles
-
The role of macrophage scavenger receptor 1 (MSR1) in inflammatory disorders and cancer.Front Immunol. 2022 Oct 17;13:1012002. doi: 10.3389/fimmu.2022.1012002. eCollection 2022. Front Immunol. 2022. PMID: 36325338 Free PMC article. Review.
-
Macrophage MSR1 promotes the formation of foamy macrophage and neuronal apoptosis after spinal cord injury.J Neuroinflammation. 2020 Feb 17;17(1):62. doi: 10.1186/s12974-020-01735-2. J Neuroinflammation. 2020. PMID: 32066456 Free PMC article.
-
RNA helicase DDX5 participates in oxLDL-induced macrophage scavenger receptor 1 expression by suppressing mRNA degradation.Exp Cell Res. 2018 May 15;366(2):114-120. doi: 10.1016/j.yexcr.2018.03.003. Epub 2018 Mar 6. Exp Cell Res. 2018. PMID: 29522752
-
Macrophage MSR1 promotes BMSC osteogenic differentiation and M2-like polarization by activating PI3K/AKT/GSK3β/β-catenin pathway.Theranostics. 2020 Jan 1;10(1):17-35. doi: 10.7150/thno.36930. eCollection 2020. Theranostics. 2020. PMID: 31903103 Free PMC article.
-
Role of macrophage scavenger receptor MSR1 in the progression of non-alcoholic steatohepatitis.Front Immunol. 2022 Dec 15;13:1050984. doi: 10.3389/fimmu.2022.1050984. eCollection 2022. Front Immunol. 2022. PMID: 36591228 Free PMC article. Review.
Cited by
-
The role of macrophage scavenger receptor 1 (MSR1) in inflammatory disorders and cancer.Front Immunol. 2022 Oct 17;13:1012002. doi: 10.3389/fimmu.2022.1012002. eCollection 2022. Front Immunol. 2022. PMID: 36325338 Free PMC article. Review.
-
Neobavaisoflavone ameliorates LPS-induced RAW264.7 cell inflammations by suppressing the activation of NF-κB and MAPKs signaling pathways.Iran J Basic Med Sci. 2022 Aug;25(8):1021-1027. doi: 10.22038/IJBMS.2022.65372.14389. Iran J Basic Med Sci. 2022. PMID: 36159335 Free PMC article.
-
Protein Arginylation Is Regulated during SARS-CoV-2 Infection.Viruses. 2023 Jan 19;15(2):290. doi: 10.3390/v15020290. Viruses. 2023. PMID: 36851505 Free PMC article.
-
Integrated transcriptome study of the tumor microenvironment for treatment response prediction in male predominant hypopharyngeal carcinoma.Nat Commun. 2023 Mar 16;14(1):1466. doi: 10.1038/s41467-023-37159-8. Nat Commun. 2023. PMID: 36928331 Free PMC article.
-
Mass spectrometry-based proteomic exploration of diverse murine macrophage cellular models.Life Sci Alliance. 2024 Nov 7;8(1):e202402760. doi: 10.26508/lsa.202402760. Print 2025 Jan. Life Sci Alliance. 2024. PMID: 39510801 Free PMC article.
References
-
- Balce DR, Li B, Allan ER, Rybicka JM, Krohn RM, Yates RM (2011) Alternative activation of macrophages by IL‐4 enhances the proteolytic capacity of their phagosomes through synergistic mechanisms. Blood 118: 4199–4208 - PubMed
-
- Balce DR, Rybicka JM, Greene CJ, Ewanchuk BW, Yates RM (2016) Ligation of FcgammaR alters phagosomal processing of protein via augmentation of NADPH oxidase activity. Traffic 17: 786–802 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

