Illuminating Biological Interactions with in Vivo Protein Footprinting
- PMID: 31025855
- PMCID: PMC6533598
- DOI: 10.1021/acs.analchem.9b00244
Illuminating Biological Interactions with in Vivo Protein Footprinting
Abstract
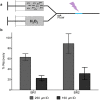
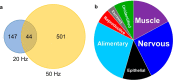
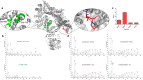
Protein footprinting coupled with mass spectrometry is being increasingly used for the study of protein interactions and conformations. The hydroxyl radical footprinting method, fast photochemical oxidation of proteins (FPOP), utilizes hydroxyl radicals to oxidatively modify solvent accessible amino acids. Here, we describe the further development of FPOP for protein structural analysis in vivo (IV-FPOP) with Caenorhabditis elegans. C. elegans, part of the nematode family, are used as model systems for many human diseases. The ability to perform structural studies in these worms would provide insight into the role of structure in disease pathogenesis. Many parameters were optimized for labeling within the worms including the microfluidic flow system and hydrogen peroxide concentration. IV-FPOP was able to modify several hundred proteins in various organs within the worms. The method successfully probed solvent accessibility similarily to in vitro FPOP, demonstrating its potential for use as a structural technique in a multiorgan system. The coupling of the method with mass spectrometry allows for amino-acid-residue-level structural information, a higher resolution than currently available in vivo methods.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


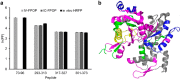
Similar articles
-
In Vivo Hydroxyl Radical Protein Footprinting for the Study of Protein Interactions in Caenorhabditis elegans.J Vis Exp. 2020 Apr 1;(158):10.3791/60910. doi: 10.3791/60910. J Vis Exp. 2020. PMID: 32310230 Free PMC article.
-
Fast Photochemical Oxidation of Proteins Coupled with Mass Spectrometry.Protein Pept Lett. 2019;26(1):27-34. doi: 10.2174/0929866526666181128124554. Protein Pept Lett. 2019. PMID: 30484399 Free PMC article. Review.
-
Validation of the Applicability of In-Cell Fast Photochemical Oxidation of Proteins across Multiple Eukaryotic Cell Lines.J Am Soc Mass Spectrom. 2020 Jul 1;31(7):1372-1379. doi: 10.1021/jasms.0c00014. Epub 2020 Mar 16. J Am Soc Mass Spectrom. 2020. PMID: 32142260
-
Chemical Penetration Enhancers Increase Hydrogen Peroxide Uptake in C. elegans for In Vivo Fast Photochemical Oxidation of Proteins.J Proteome Res. 2020 Sep 4;19(9):3708-3715. doi: 10.1021/acs.jproteome.0c00245. Epub 2020 Jun 17. J Proteome Res. 2020. PMID: 32506919 Free PMC article.
-
Mass Spectrometry-Based Fast Photochemical Oxidation of Proteins (FPOP) for Higher Order Structure Characterization.Acc Chem Res. 2018 Mar 20;51(3):736-744. doi: 10.1021/acs.accounts.7b00593. Epub 2018 Feb 16. Acc Chem Res. 2018. PMID: 29450991 Free PMC article. Review.
Cited by
-
Rapid Quantification of Peptide Oxidation Isomers From Complex Mixtures.Anal Chem. 2020 Mar 3;92(5):3834-3843. doi: 10.1021/acs.analchem.9b05268. Epub 2020 Feb 17. Anal Chem. 2020. PMID: 32039584 Free PMC article.
-
Advances in Mass Spectrometry on Membrane Proteins.Membranes (Basel). 2023 Apr 24;13(5):457. doi: 10.3390/membranes13050457. Membranes (Basel). 2023. PMID: 37233518 Free PMC article. Review.
-
Systematic Fe(II)-EDTA Method of Dose-Dependent Hydroxyl Radical Generation for Protein Oxidative Footprinting.Anal Chem. 2023 Dec 19;95(50):18316-18325. doi: 10.1021/acs.analchem.3c02319. Epub 2023 Dec 4. Anal Chem. 2023. PMID: 38049117 Free PMC article.
-
Enabling Real-Time Compensation in Fast Photochemical Oxidations of Proteins for the Determination of Protein Topography Changes.J Vis Exp. 2020 Sep 1;(163):10.3791/61580. doi: 10.3791/61580. J Vis Exp. 2020. PMID: 32955502 Free PMC article.
-
MEMBRANE PROTEIN STRUCTURES AND INTERACTIONS FROM COVALENT LABELING COUPLED WITH MASS SPECTROMETRY.Mass Spectrom Rev. 2022 Jan;41(1):51-69. doi: 10.1002/mas.21667. Epub 2020 Nov 4. Mass Spectrom Rev. 2022. PMID: 33145813 Free PMC article. Review.
References
-
- Murphy H. C. The Use of Whole Animals Versus Isolated Organs or Cell Culture in Research. Transactions of the Nebraska Academy of Sciences and Affiliated Societies 1991, 105–108.
-
- Huang R. Y.; Krystek S. R. Jr.; Felix N.; Graziano R. F.; Srinivasan M.; Pashine A.; Chen G. Hydrogen/deuterium exchange mass spectrometry and computational modeling reveal a discontinuous epitope of an antibody/TL1A Interaction. mAbs 2018, 10 (1), 95–103. 10.1080/19420862.2017.1393595. - DOI - PMC - PubMed
-
- Weis D. D., Ed. Hydrogen Exchange Mass Spectrometry of Proteins: Fundamentals, Methods, and Applications; Wiley, 2016.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

