A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses
- PMID: 31025076
- PMCID: PMC7080015
- DOI: 10.1007/s00253-019-09835-7
A TaqMan-probe-based multiplex real-time RT-qPCR for simultaneous detection of porcine enteric coronaviruses
Abstract
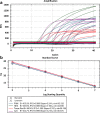
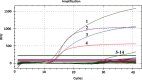
Swine enteric coronaviruses are a group of most significant pathogens causing diarrhea in piglets with similar clinical symptoms and pathological changes. To develop a simple, rapid, accurate, and high-throughput detection method for diagnosis and differential diagnosis on swine enteric coronaviruses, specific primers and probes were designed based on the highly conserved regions of transmissible gastroenteritis virus (TGEV) N, porcine epidemic diarrhea virus (PEDV) M, porcine deltacoronavirus (PDCoV) M, and porcine enteric alphacoronavirus (PEAV) N genes respectively. A TaqMan-probe-based multiplex real-time RT-qPCR assay was developed and optimized to simultaneously detect these swine enteric coronaviruses. The results showed that the limit of detection can reach as low as 10 copies in singular real-time RT-qPCR assays and 100 copies in multiplex real-time RT-qPCR assay, with all correlation coefficients (R2) at above 0.99, and the amplification efficiency at between 90 and 120%. This multiplex real-time RT-qPCR assay demonstrated high sensitivity, extreme specificity, and excellent repeatability. The multiplex real-time RT-qPCR assay was then employed to detect the swine enteric coronavirus from 354 field diarrheal samples. The results manifested that TGEV and PDCoV were the main pathogens in these samples, accompanied by co-infections. This well-established multiplex real-time RT-qPCR assay provided a rapid, efficient, specific, and sensitive tool for detection of swine enteric coronaviruses.
Keywords: Diagnosis; Multiplex real-time RT-qPCR; Swine enteric coronaviruses; TaqMan probe.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Establishment and application of a quadruplex real-time RT-qPCR assay for differentiation of TGEV, PEDV, PDCoV, and PoRVA.Microb Pathog. 2024 Jun;191:106646. doi: 10.1016/j.micpath.2024.106646. Epub 2024 Apr 16. Microb Pathog. 2024. PMID: 38631414
-
Development of a triplex RT-RAA-LFA assay for the rapid differential diagnosis of porcine epidemic diarrhea virus, porcine deltacoronavirus and transmissible gastroenteritis virus.Microb Pathog. 2024 Oct;195:106885. doi: 10.1016/j.micpath.2024.106885. Epub 2024 Aug 24. Microb Pathog. 2024. PMID: 39182857
-
Development of a multiplex RT-PCR for the detection of major diarrhoeal viruses in pig herds in China.Transbound Emerg Dis. 2020 Mar;67(2):678-685. doi: 10.1111/tbed.13385. Epub 2019 Oct 28. Transbound Emerg Dis. 2020. PMID: 31597013 Free PMC article.
-
Porcine enteric coronaviruses: an updated overview of the pathogenesis, prevalence, and diagnosis.Vet Res Commun. 2021 Sep;45(2-3):75-86. doi: 10.1007/s11259-021-09808-0. Epub 2021 Jul 12. Vet Res Commun. 2021. PMID: 34251560 Free PMC article. Review.
-
An overview of immunological and genetic methods for detecting swine coronaviruses, transmissible gastroenteritis virus, and porcine respiratory coronavirus in tissues.Adv Exp Med Biol. 1997;412:37-46. doi: 10.1007/978-1-4899-1828-4_4. Adv Exp Med Biol. 1997. PMID: 9191988 Review.
Cited by
-
Development of a Multiplex Real-Time PCR Assay for the Simultaneous Detection of Two Fungal Pathogens Causing Pneumonia.J Fungi (Basel). 2024 Aug 29;10(9):619. doi: 10.3390/jof10090619. J Fungi (Basel). 2024. PMID: 39330379 Free PMC article.
-
Detection of Four Porcine Enteric Coronaviruses Using CRISPR-Cas12a Combined with Multiplex Reverse Transcriptase Loop-Mediated Isothermal Amplification Assay.Viruses. 2022 Apr 17;14(4):833. doi: 10.3390/v14040833. Viruses. 2022. PMID: 35458562 Free PMC article.
-
Development of a Multiplex Quantitative PCR for Detecting Porcine Epidemic Diarrhea Virus, Transmissible Gastroenteritis Virus, and Porcine Deltacoronavirus Simultaneously in China.Vet Sci. 2023 Jun 18;10(6):402. doi: 10.3390/vetsci10060402. Vet Sci. 2023. PMID: 37368788 Free PMC article.
-
Development and Clinical Application of a Molecular Assay for Four Common Porcine Enteroviruses.Vet Sci. 2024 Jul 9;11(7):305. doi: 10.3390/vetsci11070305. Vet Sci. 2024. PMID: 39057989 Free PMC article.
-
Diagnostic Approach to Enteric Disorders in Pigs.Animals (Basel). 2023 Jan 18;13(3):338. doi: 10.3390/ani13030338. Animals (Basel). 2023. PMID: 36766227 Free PMC article. Review.
References
-
- Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, Faccini S, Bonilauri P, Cordioli P, Marthaler D. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. 2016;22(1):83–87. doi: 10.3201/eid2201.150544. - DOI - PMC - PubMed
-
- Chang CY, Deng MC, Wang FI, Tsai HJ, Yang CH, Chang C, Huang YL. The application of a duplex reverse transcription real-time PCR for the surveillance of porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. J Virol Methods. 2014;201:13–19. doi: 10.1016/j.jviromet.2014.01.016. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

