Subtype classification and functional annotation of L1Md retrotransposon promoters
- PMID: 31007728
- PMCID: PMC6454616
- DOI: 10.1186/s13100-019-0156-5
Subtype classification and functional annotation of L1Md retrotransposon promoters
Abstract
Background: L1Md retrotransposons are the most abundant and active transposable elements in the mouse genome. The promoters of many L1Md retrotransposons are composed of tandem repeats called monomers. The number of monomers varies between retrotransposon copies, thus making it difficult to annotate L1Md promoters. Duplication of monomers contributes to the maintenance of L1Md promoters during truncation-prone retrotranspositions, but the associated mechanism remains unclear. Since the current classification of monomers is based on limited data, a comprehensive monomer annotation is needed for supporting functional studies of L1Md promoters genome-wide.
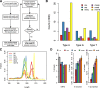
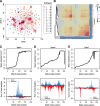
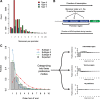
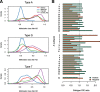
Results: We developed a pipeline for de novo monomer detection and classification. Identified monomers are further classified into subtypes based on their sequence profiles. We applied this pipeline to genome assemblies of various rodent species. A major monomer subtype of the lab mouse was also found in other Mus species, implying that such subtype has emerged in the common ancestor of involved species. We also characterized the positioning pattern of monomer subtypes within individual promoters. Our analyses indicate that the subtype composition of an L1Md promoter can be used to infer its transcriptional activity during male germ cell development.
Conclusions: We identified subtypes for all monomer types using comprehensive data, greatly expanding the spectrum of monomer variants. The analysis of monomer subtype positioning provides evidence supporting both previously proposed models of L1Md promoter expansion. The transcription silencing of L1Md promoters differs between promoter types, which supports a model involving distinct suppressive pathways rather than a universal mechanism for retrotransposon repression in gametogenesis.
Keywords: Annotation; Classification; L1Md monomer; Retrotransposon.
Conflict of interest statement
Not applicable.Not applicable.The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Composite of A and F-type 5' terminal sequences defines a subfamily of mouse LINE-1 elements.J Mol Biol. 1991 Sep 20;221(2):367-73. doi: 10.1016/0022-2836(91)80057-2. J Mol Biol. 1991. PMID: 1920423 Review.
-
Determination of a functional ancestral sequence and definition of the 5' end of A-type mouse L1 elements.J Mol Biol. 1987 Aug 20;196(4):757-67. doi: 10.1016/0022-2836(87)90402-5. J Mol Biol. 1987. PMID: 3681977
-
Switching of dominant retrotransposon silencing strategies from posttranscriptional to transcriptional mechanisms during male germ-cell development in mice.PLoS Genet. 2017 Jul 27;13(7):e1006926. doi: 10.1371/journal.pgen.1006926. eCollection 2017 Jul. PLoS Genet. 2017. PMID: 28749988 Free PMC article.
-
The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons.Mol Cell Biol. 1986 Jan;6(1):168-82. doi: 10.1128/mcb.6.1.168-182.1986. Mol Cell Biol. 1986. PMID: 3023821 Free PMC article.
-
Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline.Semin Cell Dev Biol. 2016 Nov;59:118-125. doi: 10.1016/j.semcdb.2016.03.001. Epub 2016 Mar 5. Semin Cell Dev Biol. 2016. PMID: 26957474 Free PMC article. Review.
Cited by
-
TET activity safeguards pluripotency throughout embryonic dormancy.Nat Struct Mol Biol. 2024 Oct;31(10):1625-1639. doi: 10.1038/s41594-024-01313-7. Epub 2024 May 23. Nat Struct Mol Biol. 2024. PMID: 38783076 Free PMC article.
-
Locus-specific analysis of Transposable Elements during the progression of ALS in the SOD1G93A mouse model.PLoS One. 2021 Oct 6;16(10):e0258291. doi: 10.1371/journal.pone.0258291. eCollection 2021. PLoS One. 2021. PMID: 34614020 Free PMC article.
-
Subfamily-specific differential contribution of individual monomers and the tether sequence to mouse L1 promoter activity.Mob DNA. 2022 Apr 20;13(1):13. doi: 10.1186/s13100-022-00269-z. Mob DNA. 2022. PMID: 35443687 Free PMC article.
-
Aging differentially alters the transcriptome and landscape of chromatin accessibility in the male and female mouse hippocampus.Front Mol Neurosci. 2024 Jan 22;17:1334862. doi: 10.3389/fnmol.2024.1334862. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38318533 Free PMC article.
-
Technology to the rescue: how to uncover the role of transposable elements in preimplantation development.Biochem Soc Trans. 2024 Jun 26;52(3):1349-1362. doi: 10.1042/BST20231262. Biochem Soc Trans. 2024. PMID: 38752836 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

