The priming factor CAPS1 regulates dense-core vesicle acidification by interacting with rabconnectin3β/WDR7 in neuroendocrine cells
- PMID: 31004036
- PMCID: PMC6579465
- DOI: 10.1074/jbc.RA119.007504
The priming factor CAPS1 regulates dense-core vesicle acidification by interacting with rabconnectin3β/WDR7 in neuroendocrine cells
Abstract
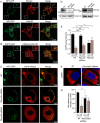
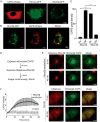
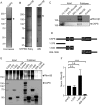
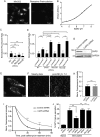
Vacuolar-type H+-ATPases (V-ATPases) contribute to pH regulation and play key roles in secretory and endocytic pathways. Dense-core vesicles (DCVs) in neuroendocrine cells are maintained at an acidic pH, which is part of the electrochemical driving force for neurotransmitter loading and is required for hormonal propeptide processing. Genetic loss of CAPS1 (aka calcium-dependent activator protein for secretion, CADPS), a vesicle-bound priming factor required for DCV exocytosis, dissipates the pH gradient across DCV membranes and reduces neurotransmitter loading. However, the basis for CAPS1 binding to DCVs and for its regulation of vesicle pH has not been determined. Here, MS analysis of CAPS1 immunoprecipitates from brain membrane fractions revealed that CAPS1 associates with a rabconnectin3 (Rbcn3) complex comprising Dmx-like 2 (DMXL2) and WD repeat domain 7 (WDR7) proteins. Using immunofluorescence microscopy, we found that Rbcn3α/DMXL2 and Rbcn3β/WDR7 colocalize with CAPS1 on DCVs in human neuroendocrine (BON) cells. The shRNA-mediated knockdown of Rbcn3β/WDR7 redistributed CAPS1 from DCVs to the cytosol, indicating that Rbcn3β/WDR7 is essential for optimal DCV localization of CAPS1. Moreover, cell-free experiments revealed direct binding of CAPS1 to Rbcn3β/WDR7, and cell assays indicated that Rbcn3β/WDR7 recruits soluble CAPS1 to membranes. As anticipated by the reported association of Rbcn3 with V-ATPase, we found that knocking down CAPS1, Rbcn3α, or Rbcn3β in neuroendocrine cells impaired rates of DCV reacidification. These findings reveal a basis for CAPS1 binding to DCVs and for CAPS1 regulation of V-ATPase activity via Rbcn3β/WDR7 interactions.
Keywords: CAPS; H+-ATPase; WDR7; cellular regulation; dense-core vesicle; exocytosis; intracellular trafficking; organellar pH homeostasis; protein-protein interaction; rabconnectin3.
© 2019 Crummy et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
Interaction of calcium-dependent activator protein for secretion 1 (CAPS1) with the class II ADP-ribosylation factor small GTPases is required for dense-core vesicle trafficking in the trans-Golgi network.J Biol Chem. 2010 Dec 3;285(49):38710-9. doi: 10.1074/jbc.M110.137414. Epub 2010 Oct 4. J Biol Chem. 2010. PMID: 20921225 Free PMC article.
-
Ca2+-dependent activator protein for secretion 1 is critical for constitutive and regulated exocytosis but not for loading of transmitters into dense core vesicles.J Biol Chem. 2007 Jul 20;282(29):21392-403. doi: 10.1074/jbc.M703699200. Epub 2007 May 31. J Biol Chem. 2007. PMID: 17540763
-
CAPS1 RNA Editing Promotes Dense Core Vesicle Exocytosis.Cell Rep. 2016 Nov 15;17(8):2004-2014. doi: 10.1016/j.celrep.2016.10.073. Cell Rep. 2016. PMID: 27851964 Free PMC article.
-
Mechanisms of exocytosis.Acta Physiol (Oxf). 2008 Feb;192(2):185-93. doi: 10.1111/j.1748-1716.2007.01803.x. Epub 2007 Nov 15. Acta Physiol (Oxf). 2008. PMID: 18005396 Review.
-
PI(4,5)P₂-binding effector proteins for vesicle exocytosis.Biochim Biophys Acta. 2015 Jun;1851(6):785-93. doi: 10.1016/j.bbalip.2014.09.017. Epub 2014 Oct 2. Biochim Biophys Acta. 2015. PMID: 25280637 Free PMC article. Review.
Cited by
-
Ion Channels and Pumps in Autophagy: A Reciprocal Relationship.Cells. 2021 Dec 14;10(12):3537. doi: 10.3390/cells10123537. Cells. 2021. PMID: 34944044 Free PMC article. Review.
-
Rabconnectin-3α/DMXL2 Is Locally Enriched at the Synaptic Ribbon of Rod Photoreceptor Synapses.Cells. 2023 Jun 19;12(12):1665. doi: 10.3390/cells12121665. Cells. 2023. PMID: 37371135 Free PMC article.
-
Localization of the Priming Factors CAPS1 and CAPS2 in Mouse Sensory Neurons Is Determined by Their N-Termini.Front Mol Neurosci. 2022 Apr 14;15:674243. doi: 10.3389/fnmol.2022.674243. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35493323 Free PMC article.
-
Trafficking of hormones and trophic factors to secretory and extracellular vesicles: a historical perspective and new hypothesis.Extracell Vesicles Circ Nucl Acids. 2023;4(4):568-587. doi: 10.20517/evcna.2023.34. Epub 2023 Nov 9. Extracell Vesicles Circ Nucl Acids. 2023. PMID: 38435713 Free PMC article.
-
RAVE and Rabconnectin-3 Complexes as Signal Dependent Regulators of Organelle Acidification.Front Cell Dev Biol. 2021 Jun 24;9:698190. doi: 10.3389/fcell.2021.698190. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34249946 Free PMC article. Review.
References
-
- Hummer B. H., de Leeuw N. F., Burns C., Chen L., Joens M. S., Hosford B., Fitzpatrick J. A. J., and Asensio C. S. (2017) HID-1 controls formation of large dense core vesicles by influencing cargo sorting and trans-Golgi network acidification. Mol. Biol. Cell 28, 3870–3880 10.1091/mbc.e17-08-0491 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

