The redox regulator sulfiredoxin forms a complex with thioredoxin domain-containing 5 protein in response to ER stress in lung cancer cells
- PMID: 31000628
- PMCID: PMC6552416
- DOI: 10.1074/jbc.RA118.005804
The redox regulator sulfiredoxin forms a complex with thioredoxin domain-containing 5 protein in response to ER stress in lung cancer cells
Abstract
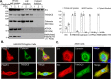
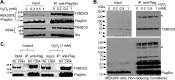
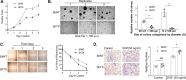
Sulfiredoxin (Srx) reduces hyperoxidized 2-cysteine-containing peroxiredoxins (Prxs) and protects cells against oxidative stress. Previous studies have shown that Srx is highly expressed in primary specimens of lung cancer patients and plays a pivotal role in lung tumorigenesis and cancer progression. However, the oncogenic mechanisms of Srx in cancer are incompletely understood. In this study, we found that Srx knockdown sensitizes lung cancer cells to endoplasmic reticulum (ER) stress-induced cell death. Through MS analysis, we determined that Srx forms a complex with the ER-resident protein thioredoxin domain-containing protein 5 (TXNDC5). Using reciprocal co-immunoprecipitation, immunofluorescence imaging, subcellular fractionation, and domain-mapping assays with site-specific mutagenesis and purified recombinant proteins, we further characterized the Srx-TXNDC5 interaction. In response to ER stress but not to oxidative stress, Srx exhibits an increased association with TXNDC5, facilitating the retention of Srx in the ER. Of note, TXNDC5 knockdown in lung cancer cells inhibited cell proliferation and repressed anchorage-independent colony formation and migration, but increased cell invasion and activation of mitogen-activated protein kinases. Using immunohistochemical staining, we demonstrate that TXNDC5 is highly expressed in patient-derived lung cancer specimens. Bioinformatics analysis of publicly available data sets revealed that those with high Srx levels have significantly shorter survival and that those with high TXNDC5 levels have longer survival. We conclude that the cellular levels of Srx and TXNDC5 may be useful as biomarkers to predict the survival of individuals with lung cancer.
Keywords: antioxidant; enzyme mechanism; oxidative stress; protein-disulfide isomerase; protein-protein interaction; proteomics; pulmonary carcinoma; sulfiredoxin; thioredoxin-domain containing 5 (TXNDC5); tumorigenesis.
© 2019 Chawsheen et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures







Similar articles
-
Cetuximab enhances cisplatin-induced endoplasmic reticulum stress-associated apoptosis in laryngeal squamous cell carcinoma cells by inhibiting expression of TXNDC5.Mol Med Rep. 2018 Mar;17(3):4767-4776. doi: 10.3892/mmr.2018.8376. Epub 2018 Jan 5. Mol Med Rep. 2018. PMID: 29328423
-
Tumor promoter-induced sulfiredoxin is required for mouse skin tumorigenesis.Carcinogenesis. 2014 May;35(5):1177-84. doi: 10.1093/carcin/bgu035. Epub 2014 Feb 6. Carcinogenesis. 2014. PMID: 24503444 Free PMC article.
-
Nuclear factor E2-related factor 2 dependent overexpression of sulfiredoxin and peroxiredoxin III in human lung cancer.Korean J Intern Med. 2011 Sep;26(3):304-13. doi: 10.3904/kjim.2011.26.3.304. Epub 2011 Sep 13. Korean J Intern Med. 2011. PMID: 22016591 Free PMC article.
-
TXNDC5, a newly discovered disulfide isomerase with a key role in cell physiology and pathology.Int J Mol Sci. 2014 Dec 17;15(12):23501-18. doi: 10.3390/ijms151223501. Int J Mol Sci. 2014. PMID: 25526565 Free PMC article. Review.
-
The role and mechanism of TXNDC5 in diseases.Eur J Med Res. 2022 Aug 8;27(1):145. doi: 10.1186/s40001-022-00770-4. Eur J Med Res. 2022. PMID: 35934705 Free PMC article. Review.
Cited by
-
TXNDC5 Plays a Crucial Role in Regulating Endoplasmic Reticulum Activity through Different ER Stress Signaling Pathways in Hepatic Cells.Int J Mol Sci. 2024 Jun 28;25(13):7128. doi: 10.3390/ijms25137128. Int J Mol Sci. 2024. PMID: 39000233 Free PMC article.
-
Endoplasmic Reticulum Protein TXNDC5 Interacts with PRDX6 and HSPA9 to Regulate Glutathione Metabolism and Lipid Peroxidation in the Hepatic AML12 Cell Line.Int J Mol Sci. 2023 Dec 5;24(24):17131. doi: 10.3390/ijms242417131. Int J Mol Sci. 2023. PMID: 38138960 Free PMC article.
-
ASF1B promotes cervical cancer progression through stabilization of CDK9.Cell Death Dis. 2020 Aug 26;11(8):705. doi: 10.1038/s41419-020-02872-5. Cell Death Dis. 2020. PMID: 32848135 Free PMC article.
-
Squalene Loaded Nanoparticles Effectively Protect Hepatic AML12 Cell Lines against Oxidative and Endoplasmic Reticulum Stress in a TXNDC5-Dependent Way.Antioxidants (Basel). 2022 Mar 18;11(3):581. doi: 10.3390/antiox11030581. Antioxidants (Basel). 2022. PMID: 35326231 Free PMC article.
-
METTL3 potentiates progression of cervical cancer by suppressing ER stress via regulating m6A modification of TXNDC5 mRNA.Oncogene. 2022 Sep;41(39):4420-4432. doi: 10.1038/s41388-022-02435-2. Epub 2022 Aug 20. Oncogene. 2022. PMID: 35987795
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

