Complex Membrane Remodeling during Virion Assembly of the 30,000-Year-Old Mollivirus Sibericum
- PMID: 30996095
- PMCID: PMC6580955
- DOI: 10.1128/JVI.00388-19
Complex Membrane Remodeling during Virion Assembly of the 30,000-Year-Old Mollivirus Sibericum
Abstract
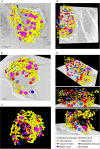
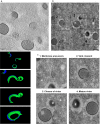
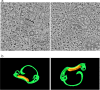
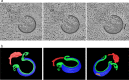
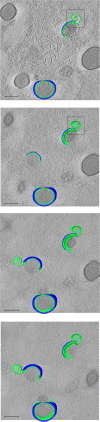
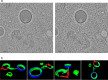
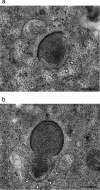
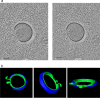
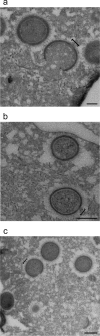
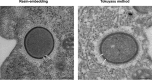
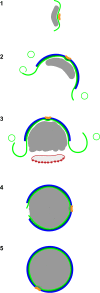
Cellular membranes ensure functional compartmentalization by dynamic fusion-fission remodeling and are often targeted by viruses during entry, replication, assembly, and egress. Nucleocytoplasmic large DNA viruses (NCLDVs) can recruit host-derived open membrane precursors to form their inner viral membrane. Using complementary three-dimensional (3D)-electron microscopy techniques, including focused-ion beam scanning electron microscopy and electron tomography, we show that the giant Mollivirus sibericum utilizes the same strategy but also displays unique features. Indeed, assembly is specifically triggered by an open cisterna with a flat pole in its center and open curling ends that grow by recruitment of vesicles never reported for NCLDVs. These vesicles, abundant in the viral factory (VF), are initially closed but open once in close proximity to the open curling ends of the growing viral membrane. The flat pole appears to play a central role during the entire virus assembly process. While additional capsid layers are assembled from it, it also shapes the growing cisterna into immature crescent-like virions and is located opposite to the membrane elongation and closure sites, thereby providing virions with a polarity. In the VF, DNA-associated filaments are abundant, and DNA is packed within virions prior to particle closure. Altogether, our results highlight the complexity of the interaction between giant viruses and their host. Mollivirus assembly relies on the general strategy of vesicle recruitment, opening, and shaping by capsid layers similar to all NCLDVs studied until now. However, the specific features of its assembly suggest that the molecular mechanisms for cellular membrane remodeling and persistence are unique.IMPORTANCE Since the first giant virus Mimivirus was identified, other giant representatives are isolated regularly around the world and appear to be unique in several aspects. They belong to at least four viral families, and the ways they interact with their hosts remain poorly understood. We focused on Mollivirus sibericum, the sole representative of "Molliviridae," which was isolated from a 30,000-year-old permafrost sample and exhibits spherical virions of complex composition. In particular, we show that (i) assembly is initiated by a unique structure containing a flat pole positioned at the center of an open cisterna, (ii) core packing involves another cisterna-like element seemingly pushing core proteins into particles being assembled, and (iii) specific filamentous structures contain the viral genome before packaging. Altogether, our findings increase our understanding of how complex giant viruses interact with their host and provide the foundation for future studies to elucidate the molecular mechanisms of Mollivirus assembly.
Keywords: Mollivirus sibericum; electron tomography; focused-ion beam scanning electron microscopy; giant viruses; membrane remodeling; nucleocytoplasmic large DNA viruses; viral factory; virus assembly.
Copyright © 2019 American Society for Microbiology.
Figures



Similar articles
-
Characterization of Mollivirus kamchatka, the First Modern Representative of the Proposed Molliviridae Family of Giant Viruses.J Virol. 2020 Mar 31;94(8):e01997-19. doi: 10.1128/JVI.01997-19. Print 2020 Mar 31. J Virol. 2020. PMID: 31996429 Free PMC article.
-
In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba.Proc Natl Acad Sci U S A. 2015 Sep 22;112(38):E5327-35. doi: 10.1073/pnas.1510795112. Epub 2015 Sep 8. Proc Natl Acad Sci U S A. 2015. PMID: 26351664 Free PMC article.
-
The Large Marseillevirus Explores Different Entry Pathways by Forming Giant Infectious Vesicles.J Virol. 2016 May 12;90(11):5246-55. doi: 10.1128/JVI.00177-16. Print 2016 Jun 1. J Virol. 2016. PMID: 26984730 Free PMC article.
-
Infection cycles of large DNA viruses: emerging themes and underlying questions.Virology. 2014 Oct;466-467:3-14. doi: 10.1016/j.virol.2014.05.037. Epub 2014 Jul 2. Virology. 2014. PMID: 24996494 Review.
-
Giant Viruses of Amoebae: A Journey Through Innovative Research and Paradigm Changes.Annu Rev Virol. 2017 Sep 29;4(1):61-85. doi: 10.1146/annurev-virology-101416-041816. Epub 2017 Jul 31. Annu Rev Virol. 2017. PMID: 28759330 Review.
Cited by
-
Cytopathic effects in Mimivirus infection: understanding the kinetics of virus-cell interaction.Mem Inst Oswaldo Cruz. 2024 Jul 22;119:e230186. doi: 10.1590/0074-02760230186. eCollection 2024. Mem Inst Oswaldo Cruz. 2024. PMID: 39045993 Free PMC article.
-
Evolution of giant pandoravirus revealed by CRISPR/Cas9.Nat Commun. 2023 Jan 26;14(1):428. doi: 10.1038/s41467-023-36145-4. Nat Commun. 2023. PMID: 36702819 Free PMC article.
-
Unpicking the Secrets of African Swine Fever Viral Replication Sites.Viruses. 2021 Jan 8;13(1):77. doi: 10.3390/v13010077. Viruses. 2021. PMID: 33429879 Free PMC article.
-
Megaviruses contain various genes encoding for eukaryotic vesicle trafficking factors.Traffic. 2022 Aug;23(8):414-425. doi: 10.1111/tra.12860. Epub 2022 Jun 28. Traffic. 2022. PMID: 35701729 Free PMC article.
-
Giant virus biology and diversity in the era of genome-resolved metagenomics.Nat Rev Microbiol. 2022 Dec;20(12):721-736. doi: 10.1038/s41579-022-00754-5. Epub 2022 Jul 28. Nat Rev Microbiol. 2022. PMID: 35902763 Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

