Regulation of signaling mediated by nucleic acid sensors for innate interferon-mediated responses during viral infection
- PMID: 30985869
- PMCID: PMC7110195
- DOI: 10.1093/intimm/dxz034
Regulation of signaling mediated by nucleic acid sensors for innate interferon-mediated responses during viral infection
Abstract
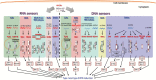
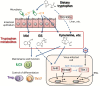
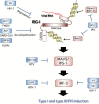
Type I and type III interferons are important anti-viral cytokines that are massively induced during viral infection. This dynamic process is regulated by many executors and regulators for efficient eradication of invading viruses and protection from harmful, excessive responses. An array of innate sensors recognizes virus-derived nucleic acids to activate their downstream signaling to evoke cytokine responses including interferons. In particular, a cytoplasmic RNA sensor RIG-I (retinoic acid-inducible gene I) is involved in the detection of multiple types of not only RNA viruses but also DNA viruses. Accumulating findings have revealed that activation of nucleic acid sensors and the related signaling mediators is regulated on the basis of post-translational modification such as ubiquitination, phosphorylation and ADP-ribosylation. In addition, long non-coding RNAs (lncRNAs) have been implicated as a new class of regulators in innate signaling. A comprehensive understanding of the regulatory mechanisms of innate sensor activation and its signaling in host-virus interaction will provide a better therapeutic strategy to efficiently control viral infection and maintain immune homeostasis.
Keywords: RIG-I; interferon; pattern-recognition receptors; signal transduction; virus infection.
© The Japanese Society for Immunology. 2019. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

Similar articles
-
Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors.Cell Mol Immunol. 2021 Mar;18(3):539-555. doi: 10.1038/s41423-020-00602-7. Epub 2021 Jan 18. Cell Mol Immunol. 2021. PMID: 33462384 Free PMC article. Review.
-
Post-translational regulation of antiviral innate signaling.Eur J Immunol. 2017 Sep;47(9):1414-1426. doi: 10.1002/eji.201746959. Epub 2017 Aug 14. Eur J Immunol. 2017. PMID: 28744851 Free PMC article. Review.
-
Camouflage and interception: how pathogens evade detection by intracellular nucleic acid sensors.Immunology. 2019 Mar;156(3):217-227. doi: 10.1111/imm.13030. Epub 2018 Dec 18. Immunology. 2019. PMID: 30499584 Free PMC article. Review.
-
Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity.Pharmacol Ther. 2009 Nov;124(2):219-34. doi: 10.1016/j.pharmthera.2009.06.012. Epub 2009 Jul 15. Pharmacol Ther. 2009. PMID: 19615405 Free PMC article. Review.
-
Recognition of viral nucleic acids in innate immunity.Rev Med Virol. 2010 Jan;20(1):4-22. doi: 10.1002/rmv.633. Rev Med Virol. 2010. PMID: 20041442 Review.
Cited by
-
Identification of Nonsynonymous SNPs in Immune-Related Genes Associated with Pneumonia Severity in Pigs.Genes (Basel). 2024 Aug 21;15(8):1103. doi: 10.3390/genes15081103. Genes (Basel). 2024. PMID: 39202462 Free PMC article.
-
Characterization of SARS-CoV-2 Evasion: Interferon Pathway and Therapeutic Options.Viruses. 2022 Jun 8;14(6):1247. doi: 10.3390/v14061247. Viruses. 2022. PMID: 35746718 Free PMC article. Review.
-
Dual Effect of Organogermanium Compound THGP on RIG-I-Mediated Viral Sensing and Viral Replication during Influenza a Virus Infection.Viruses. 2021 Aug 24;13(9):1674. doi: 10.3390/v13091674. Viruses. 2021. PMID: 34578256 Free PMC article.
-
ISG15: A link between innate immune signaling, DNA replication, and genome stability.Bioessays. 2023 Jul;45(7):e2300042. doi: 10.1002/bies.202300042. Epub 2023 May 5. Bioessays. 2023. PMID: 37147792 Free PMC article. Review.
-
Plant-Derived Food Grade Substances (PDFGS) Active Against Respiratory Viruses: A Systematic Review of Non-clinical Studies.Front Nutr. 2021 Feb 9;8:606782. doi: 10.3389/fnut.2021.606782. eCollection 2021. Front Nutr. 2021. PMID: 33634160 Free PMC article.
References
-
- Nagano Y. and Kojima Y. 1954. [Immunizing property of vaccinia virus inactivated by ultraviolets rays]. C. R. Seances Soc. Biol. Fil. 148:1700. - PubMed
-
- Isaacs A. and Lindenmann J. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B. Biol. Sci. 147:258. - PubMed
-
- Kumar H., Kawai T. and Akira S. 2011. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30:16. - PubMed
-
- Alexopoulou L., Holt A. C., Medzhitov R. and Flavell R. A. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413:732. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

