Diversity of the microbial community and cultivable protease-producing bacteria in the sediments of the Bohai Sea, Yellow Sea and South China Sea
- PMID: 30973915
- PMCID: PMC6459509
- DOI: 10.1371/journal.pone.0215328
Diversity of the microbial community and cultivable protease-producing bacteria in the sediments of the Bohai Sea, Yellow Sea and South China Sea
Abstract
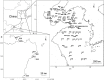
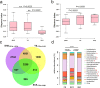
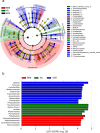
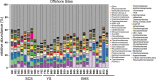
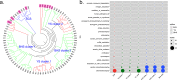
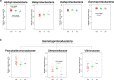
The nitrogen (N) cycle is closely related to the stability of marine ecosystems. Microbial communities have been directly linked to marine N-cycling processes. However, systematic research on the bacterial community composition and diversity involved in N cycles in different seas is lacking. In this study, microbial diversity in the Bohai Sea (BHS), Yellow Sea (YS) and South China Sea (SCS) was surveyed by targeting the hypervariable V4 regions of the 16S rRNA gene using next-generation sequencing (NGS) technology. A total of 2,505,721 clean reads and 15,307 operational taxonomic units (OTUs) were obtained from 86 sediment samples from the three studied China seas. LEfSe analysis demonstrated that the SCS had more abundant microbial taxa than the BHS and YS. Diversity indices demonstrated that Proteobacteria and Planctomycetes were the dominant phyla in all three China seas. Canonical correspondence analysis (CCA) indicated that pH (P = 0.034) was the principal determining factors, while the organic matter content, depth and temperature had a minor correlated with the variations in sedimentary microbial community distribution. Cluster and functional analyses of microbial communities showed that chemoheterotrophic and aerobic chemoheterotrophic microorganisms widely exist in these three seas. Further research found that the cultivable protease-producing bacteria were mainly affiliated with the phyla Proteobacteria, Firmicutes and Bacteroidetes. It was very clear that Pseudoalteromonadaceae possessed the highest relative abundance in the three sea areas. The predominant protease-producing genera were Pseudoalteromonas and Bacillus. These results shed light on the differences in bacterial community composition, especially protease-producing bacteria, in these three China seas.
Conflict of interest statement
The authors have the following interests: Ming Chen, Xinwu Guo and Jun Wang are employee of Sanway Gene Technology. There are no patents, products in development or marketed products to declare. This does not alter our adherence to all the PLOS ONE policies on sharing data and materials.
Figures

Similar articles
-
Nutrients and heavy metals mediate the distribution of microbial community in the marine sediments of the Bohai Sea, China.Environ Pollut. 2019 Dec;255(Pt 1):113069. doi: 10.1016/j.envpol.2019.113069. Epub 2019 Aug 24. Environ Pollut. 2019. PMID: 31541809
-
Bacterial community composition of South China Sea sediments through pyrosequencing-based analysis of 16S rRNA genes.PLoS One. 2013 Oct 21;8(10):e78501. doi: 10.1371/journal.pone.0078501. eCollection 2013. PLoS One. 2013. PMID: 24205246 Free PMC article.
-
Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China sea.Microb Ecol. 2009 Oct;58(3):582-90. doi: 10.1007/s00248-009-9506-z. Epub 2009 Mar 20. Microb Ecol. 2009. PMID: 19301066
-
Under the sea: microbial life in volcanic oceanic crust.Nat Rev Microbiol. 2011 Sep 6;9(10):703-12. doi: 10.1038/nrmicro2647. Nat Rev Microbiol. 2011. PMID: 21894169 Review.
-
Microbial Diversity in the Indian Ocean Sediments: An Insight into the Distribution and Associated Factors.Curr Microbiol. 2022 Feb 23;79(4):115. doi: 10.1007/s00284-022-02801-z. Curr Microbiol. 2022. PMID: 35195780 Review.
Cited by
-
Gut Microbial Composition and Diversity in Four Ophiuroid Species: Divergence Between Suspension Feeder and Scavenger and Their Symbiotic Microbes.Front Microbiol. 2021 Mar 19;12:645070. doi: 10.3389/fmicb.2021.645070. eCollection 2021. Front Microbiol. 2021. PMID: 33815331 Free PMC article.
-
The interplay between host-specificity and habitat-filtering influences sea cucumber microbiota across an environmental gradient of pollution.Environ Microbiome. 2024 Oct 13;19(1):74. doi: 10.1186/s40793-024-00620-2. Environ Microbiome. 2024. PMID: 39397007 Free PMC article.
-
Analysis of the metals and metalloids concentrations and of the bacterial population in sediments of the Red River Delta, Vietnam.Front Microbiol. 2024 Jun 5;15:1394998. doi: 10.3389/fmicb.2024.1394998. eCollection 2024. Front Microbiol. 2024. PMID: 38933021 Free PMC article.
-
Comparative metagenomics reveals the microbial diversity and metabolic potentials in the sediments and surrounding seawaters of Qinhuangdao mariculture area.PLoS One. 2020 Jun 4;15(6):e0234128. doi: 10.1371/journal.pone.0234128. eCollection 2020. PLoS One. 2020. PMID: 32497143 Free PMC article.
-
Estuarine mangrove niches select cultivable heterotrophic diazotrophs with diverse metabolic potentials-a prospective cross-dialog for functional diazotrophy.Front Microbiol. 2024 May 24;15:1324188. doi: 10.3389/fmicb.2024.1324188. eCollection 2024. Front Microbiol. 2024. PMID: 38873137 Free PMC article.
References
-
- Jorgensen SL, Hannisdal B, Lanzen A, Baumberger T, Flesland K, Fonseca R, et al. Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge. Proc Natl Acad Sci U S A. 2012;109(42):E2846–E55. 10.1073/pnas.1207574109 WOS:000310515800004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

