The effect of HMGB1 on the clinicopathological and prognostic features of cervical cancer
- PMID: 30962261
- PMCID: PMC6499451
- DOI: 10.1042/BSR20181016
The effect of HMGB1 on the clinicopathological and prognostic features of cervical cancer
Erratum in
-
Correction: The effect of HMGB1 on the clinicopathological and prognostic features of cervical cancer.Biosci Rep. 2020 Jun 26;40(6):BSR-20181016_COR. doi: 10.1042/BSR-20181016_COR. Biosci Rep. 2020. PMID: 32556119 Free PMC article. No abstract available.
Abstract
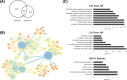
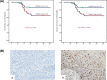
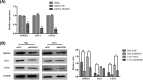
Cervical cancer is the third leading cause of cancer death among women in less-developed regions. Because of the poor survivorship of patients with advanced disease, finding new biomarkers for prognostic prediction is of great importance. In the current study, mRNA datasets (GSE9750 and GSE63514) were retrieved from Gene Expression Omnibus and was used to identify differentially expressed genes. The underlying molecular mechanisms associated with high-mobility group box 1 protein (HMGB1) were investigated using bioinformatics analysis. Immunohistochemical analysis of HMGB1 was performed on 239 cases of cervical cancer samples to investigate its possible correlation with clinicopathological characteristics and outcomes. A preliminary validation has been made to explore the possible correlation factors with HMGB1 that promote migration of cervical cancer cells. Bioinformatics analysis showed that adherens junction was significant for both P-value and enrichment scores, which was consistent with the clinical study. The underlying molecular mechanisms might be the interaction among HMGB1, RAC1, and CDC42. HMGB1 expression was significantly associated with tumor size, parametrial infiltration, the depth of cervical stromal invasion, and FIGO stage (P=0.003, 0.019, 0.013, and 0.003, respectively). FIGO stage, lymph mode metastasis, and HMGB1 expression were independent predictors of a poorer prognosis of patients with cervical cancer. Knockdown of HMGB1 inhibits migration of Siha and C33A cells in vitro Western blot and quantitative real-time PCR (qRT-PCR) showed that the expression of RAC1 and CDC42 was positively correlated with HMGB1. HMGB1 is a useful prognostic indicator and a potential biomarker of cervical cancer. RAC1 and CDC42 may be involved in the progression of cervical cancer migration induced by HMGB1.
Keywords: HMGB1; bioinformatics analysis; cervical cancer; prognosis.
© 2019 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
High-mobility group box 1 is overexpressed in cervical carcinoma and promotes cell invasion and migration in vitro.Oncol Rep. 2017 Feb;37(2):831-840. doi: 10.3892/or.2016.5317. Epub 2016 Dec 14. Oncol Rep. 2017. PMID: 28000879
-
High-mobility group box 1 expression and lymph node metastasis in intrahepatic cholangiocarcinoma.World J Gastroenterol. 2015 Mar 21;21(11):3256-65. doi: 10.3748/wjg.v21.i11.3256. World J Gastroenterol. 2015. PMID: 25805932 Free PMC article.
-
MiR-142 inhibits the development of cervical cancer by targeting HMGB1.Oncotarget. 2017 Jan 17;8(3):4001-4007. doi: 10.18632/oncotarget.13136. Oncotarget. 2017. PMID: 27829233 Free PMC article.
-
HMGB1 overexpression as a prognostic factor for survival in cancer: a meta-analysis and systematic review.Oncotarget. 2016 Aug 2;7(31):50417-50427. doi: 10.18632/oncotarget.10413. Oncotarget. 2016. PMID: 27391431 Free PMC article. Review.
-
HMGB1: an overview of its versatile roles in the pathogenesis of colorectal cancer.Cell Oncol (Dordr). 2020 Apr;43(2):177-193. doi: 10.1007/s13402-019-00477-5. Epub 2019 Nov 1. Cell Oncol (Dordr). 2020. PMID: 31677065 Review.
Cited by
-
High Mobility Group Box 1 in Human Cancer.Cells. 2020 Jul 10;9(7):1664. doi: 10.3390/cells9071664. Cells. 2020. PMID: 32664328 Free PMC article. Review.
-
HMGB1 as a therapeutic target in disease.J Cell Physiol. 2021 May;236(5):3406-3419. doi: 10.1002/jcp.30125. Epub 2020 Oct 26. J Cell Physiol. 2021. PMID: 33107103 Free PMC article. Review.
-
miR-22 alleviates sepsis-induced acute kidney injury via targeting the HMGB1/TLR4/NF-κB signaling pathway.Int Urol Nephrol. 2023 Feb;55(2):409-421. doi: 10.1007/s11255-022-03321-2. Epub 2022 Aug 12. Int Urol Nephrol. 2023. PMID: 35960478 Free PMC article.
-
HMGB1 overexpression promotes a malignant phenotype and radioresistance in ESCC.J Cancer. 2022 May 21;13(9):2717-2726. doi: 10.7150/jca.73761. eCollection 2022. J Cancer. 2022. PMID: 35812184 Free PMC article.
-
High Mobility Group Box 1: Biological Functions and Relevance in Oxidative Stress Related Chronic Diseases.Cells. 2022 Mar 1;11(5):849. doi: 10.3390/cells11050849. Cells. 2022. PMID: 35269471 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

