Evolution of folate biosynthesis and metabolism across algae and land plant lineages
- PMID: 30952916
- PMCID: PMC6451014
- DOI: 10.1038/s41598-019-42146-5
Evolution of folate biosynthesis and metabolism across algae and land plant lineages
Abstract
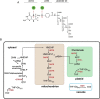
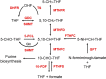
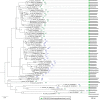
Tetrahydrofolate and its derivatives, commonly known as folates, are essential for almost all living organisms. Besides acting as one-carbon donors and acceptors in reactions producing various important biomolecules such as nucleic and amino acids, as well as pantothenate, they also supply one-carbon units for methylation reactions. Plants along with bacteria, yeast and fungi synthesize folates de novo and therefore constitute a very important dietary source of folates for animals. All the major steps of folate biosynthesis and metabolism have been identified but only few have been genetically characterized in a handful of model plant species. The possible differences in the folate pathway between various plant and algal species have never been explored. In this study we present a comprehensive comparative study of folate biosynthesis and metabolism of all major land plant lineages as well as green and red algae. The study identifies new features of plant folate metabolism that might open new directions to folate research in plants.
Conflict of interest statement
The authors declare no competing interests.
Figures




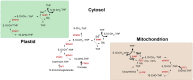
Similar articles
-
Classification, naming and evolutionary history of glycosyltransferases from sequenced green and red algal genomes.PLoS One. 2013 Oct 16;8(10):e76511. doi: 10.1371/journal.pone.0076511. eCollection 2013. PLoS One. 2013. PMID: 24146880 Free PMC article.
-
Folates and one-carbon metabolism in plants and fungi.Phytochemistry. 1997 Jun;45(3):437-52. doi: 10.1016/s0031-9422(96)00833-3. Phytochemistry. 1997. PMID: 9190084 Review.
-
Folates in Plants: Research Advances and Progress in Crop Biofortification.Front Chem. 2017 Mar 29;5:21. doi: 10.3389/fchem.2017.00021. eCollection 2017. Front Chem. 2017. PMID: 28424769 Free PMC article. Review.
-
Folate biosynthesis, turnover, and transport in plants.Annu Rev Plant Biol. 2011;62:105-25. doi: 10.1146/annurev-arplant-042110-103819. Annu Rev Plant Biol. 2011. PMID: 21275646 Review.
-
Embryophyte stress signaling evolved in the algal progenitors of land plants.Proc Natl Acad Sci U S A. 2018 Apr 10;115(15):E3471-E3480. doi: 10.1073/pnas.1719230115. Epub 2018 Mar 26. Proc Natl Acad Sci U S A. 2018. PMID: 29581286 Free PMC article.
Cited by
-
Influence of Sulfonamide Contamination Derived from Veterinary Antibiotics on Plant Growth and Development.Antibiotics (Basel). 2020 Jul 28;9(8):456. doi: 10.3390/antibiotics9080456. Antibiotics (Basel). 2020. PMID: 32731577 Free PMC article.
-
Challenging microalgal vitamins for human health.Microb Cell Fact. 2020 Nov 2;19(1):201. doi: 10.1186/s12934-020-01459-1. Microb Cell Fact. 2020. PMID: 33138823 Free PMC article. Review.
-
Dual transcriptome based reconstruction of Salmonella-human integrated metabolic network to screen potential drug targets.PLoS One. 2022 May 24;17(5):e0268889. doi: 10.1371/journal.pone.0268889. eCollection 2022. PLoS One. 2022. PMID: 35609089 Free PMC article.
-
Genome-Wide Association Study of Seed Folate Content in Common Bean.Front Plant Sci. 2021 Aug 31;12:696423. doi: 10.3389/fpls.2021.696423. eCollection 2021. Front Plant Sci. 2021. PMID: 34531882 Free PMC article.
-
Effect of Amino Acids on Folates Accumulation in Wheat Seedlings during Germination under Red Light Radiation.Molecules. 2022 Oct 13;27(20):6868. doi: 10.3390/molecules27206868. Molecules. 2022. PMID: 36296459 Free PMC article.
References
-
- James TY, et al. The pab1 gene of Coprinus cinereus encodes a bifunctional protein for paraaminobenzoic acid (PABA) synthesis: implications for the evolution of fused PABA synthases. Journal of Basic Microbiology. 2002;42:91–103. doi: 10.1002/1521-4028(200205)42:2<91::AID-JOBM91>3.0.CO;2-8. - DOI - PubMed
-
- Basset GJ, et al. Folate synthesis in plants: the p-aminobenzoate branch is initiated by a bifunctional PabAPabB protein that is targeted to plastids. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1496–1501. doi: 10.1073/pnas.0308331100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

