The mRNA export adaptor Yra1 contributes to DNA double-strand break repair through its C-box domain
- PMID: 30951522
- PMCID: PMC6450643
- DOI: 10.1371/journal.pone.0206336
The mRNA export adaptor Yra1 contributes to DNA double-strand break repair through its C-box domain
Abstract
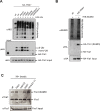
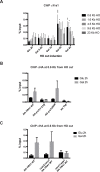
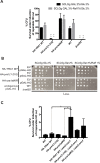
Yra1 is an mRNA export adaptor involved in mRNA biogenesis and export in S. cerevisiae. Yra1 overexpression was recently shown to promote accumulation of DNA:RNA hybrids favoring DNA double strand breaks (DSB), cell senescence and telomere shortening, via an unknown mechanism. Yra1 was also identified at an HO-induced DSB and Yra1 depletion causes defects in DSB repair. Previous work from our laboratory showed that Yra1 ubiquitination by Tom1 is important for mRNA export. Here, we found that Yra1 is also ubiquitinated by the SUMO-targeted ubiquitin ligases Slx5-Slx8 implicated in the interaction of irreparable DSB with nuclear pores. We further show that Yra1 binds an HO-induced irreparable DSB in a process dependent on resection. Importantly, a Yra1 mutant lacking the evolutionarily conserved C-box is not recruited to an HO-induced irreparable DSB and becomes lethal under DSB induction in a HO-cut reparable system. Together, the data provide evidence that Yra1 plays a crucial role in DSB repair via homologous recombination. While Yra1 sumoylation and/or ubiquitination are dispensable, the Yra1 C-box region is essential in this process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
The functional complexity of the RNA-binding protein Yra1: mRNA biogenesis, genome stability and DSB repair.Curr Genet. 2020 Feb;66(1):63-71. doi: 10.1007/s00294-019-01011-8. Epub 2019 Jul 10. Curr Genet. 2020. PMID: 31292684 Review.
-
Disruption of SUMO-targeted ubiquitin ligases Slx5-Slx8/RNF4 alters RecQ-like helicase Sgs1/BLM localization in yeast and human cells.DNA Repair (Amst). 2015 Feb;26:1-14. doi: 10.1016/j.dnarep.2014.12.004. Epub 2014 Dec 26. DNA Repair (Amst). 2015. PMID: 25588990 Free PMC article.
-
Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export.Genes Dev. 2010 Sep 1;24(17):1927-38. doi: 10.1101/gad.583310. Genes Dev. 2010. PMID: 20810649 Free PMC article.
-
Proteomic identification of histone post-translational modifications and proteins enriched at a DNA double-strand break.Nucleic Acids Res. 2017 Nov 2;45(19):10923-10940. doi: 10.1093/nar/gkx844. Nucleic Acids Res. 2017. PMID: 29036368 Free PMC article.
-
CtIP/Ctp1/Sae2, molecular form fit for function.DNA Repair (Amst). 2017 Aug;56:109-117. doi: 10.1016/j.dnarep.2017.06.013. Epub 2017 Jun 9. DNA Repair (Amst). 2017. PMID: 28623092 Free PMC article. Review.
Cited by
-
The functional complexity of the RNA-binding protein Yra1: mRNA biogenesis, genome stability and DSB repair.Curr Genet. 2020 Feb;66(1):63-71. doi: 10.1007/s00294-019-01011-8. Epub 2019 Jul 10. Curr Genet. 2020. PMID: 31292684 Review.
-
CRISPR/Cas9-mediated point mutations improve α-amylase secretion in Saccharomyces cerevisiae.FEMS Yeast Res. 2022 Jul 15;22(1):foac033. doi: 10.1093/femsyr/foac033. FEMS Yeast Res. 2022. PMID: 35776981 Free PMC article.
-
Gross Chromosomal Rearrangement at Centromeres.Biomolecules. 2023 Dec 24;14(1):28. doi: 10.3390/biom14010028. Biomolecules. 2023. PMID: 38254628 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

