The human adenovirus type 5 E1B 55kDa protein interacts with RNA promoting timely DNA replication and viral late mRNA metabolism
- PMID: 30943256
- PMCID: PMC6447194
- DOI: 10.1371/journal.pone.0214882
The human adenovirus type 5 E1B 55kDa protein interacts with RNA promoting timely DNA replication and viral late mRNA metabolism
Abstract
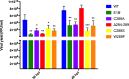
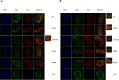
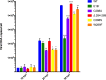
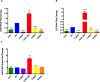
The E1B 55kDa produced by human adenovirus type 5 is a multifunctional protein that participates in the regulation of several steps during the viral replication cycle. Previous studies suggest this protein plays an important role in postranscriptional regulation of viral and cellular gene expression, as it is required for the selective accumulation of maximal levels of viral late mRNA in the cytoplasm of the infected cell; however the molecular mechanisms that are altered or regulated by this protein have not been elucidated. A ribonucleoprotein motif that could implicate the direct interaction of the protein with RNA was initially predicted and tested in vitro, but the interaction with RNA could not be detected in infected cells, suggesting the interaction may be weak or transient. Here it was determined that the E1B 55kDa interacts with RNA in the context of the viral infection in non-transformed human cells, and its contribution to the adenovirus replication cycle was evaluated. Using recombinant adenoviruses with amino acid substitutions or a deletion in the ribonucleoprotein motif the interaction of E1B 55kDa with RNA was found to correlate with timely and efficient viral DNA replication and viral late mRNA accumulation and splicing.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




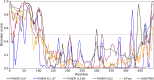

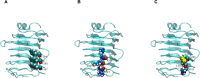
Similar articles
-
Normal human cell proteins that interact with the adenovirus type 5 E1B 55kDa protein.Virology. 2017 Apr;504:12-24. doi: 10.1016/j.virol.2017.01.013. Epub 2017 Jan 27. Virology. 2017. PMID: 28135605 Free PMC article.
-
Diverse roles for E4orf3 at late times of infection revealed in an E1B 55-kilodalton protein mutant background.J Virol. 2004 Sep;78(18):9924-35. doi: 10.1128/JVI.78.18.9924-9935.2004. J Virol. 2004. PMID: 15331726 Free PMC article.
-
E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs.J Virol. 1998 Oct;72(10):7960-71. doi: 10.1128/JVI.72.10.7960-7971.1998. J Virol. 1998. PMID: 9733834 Free PMC article.
-
The biology of the adenovirus E1B 55K protein.FEBS Lett. 2019 Dec;593(24):3504-3517. doi: 10.1002/1873-3468.13694. Epub 2019 Dec 8. FEBS Lett. 2019. PMID: 31769868 Review.
-
Genetically modified adenoviruses against gliomas: from bench to bedside.Neurology. 2004 Aug 10;63(3):418-26. doi: 10.1212/01.wnl.0000133302.15022.7f. Neurology. 2004. PMID: 15304571 Review.
Cited by
-
Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif.Viruses. 2023 Nov 30;15(12):2356. doi: 10.3390/v15122356. Viruses. 2023. PMID: 38140597 Free PMC article.
-
Global Transcriptome Analyses of Cellular and Viral mRNAs during HAdV-C5 Infection Highlight New Aspects of Viral mRNA Biogenesis and Cytoplasmic Viral mRNA Accumulations.Viruses. 2022 Nov 1;14(11):2428. doi: 10.3390/v14112428. Viruses. 2022. PMID: 36366526 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

