Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review)
- PMID: 30942418
- PMCID: PMC6471501
- DOI: 10.3892/mmr.2019.10111
Regulation of histone arginine methylation/demethylation by methylase and demethylase (Review)
Abstract
Histone arginine methylation is a universal post‑translational modification that has been implicated in multiple cellular and sub‑cellular processes, including pre‑mRNA splicing, DNA damage signaling, mRNA translation, cell signaling and cell death. Despite these important roles, the understanding of its regulation with respect to certain other modifications, such as phosphorylation and acetylation, is very poor. Thus far, few histone arginine demethylases have been identified in mammalian cells, compared with nine protein arginine methyltransferases (PRMTs) that have been reported. Studies have reported that aberrant histone arginine methylation is strongly associated with carcinogenesis and metastasis. This increases the requirement for understanding the regulation of histone arginine demethylation. The present review summarizes the published studies and provides further insights into histone arginine methylases and demethylases.
Figures
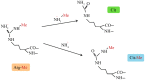

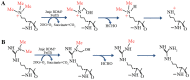
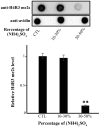
Similar articles
-
[Advances in histone methyltransferases and histone demethylases].Ai Zheng. 2008 Oct;27(10):1018-25. Ai Zheng. 2008. PMID: 18851779 Review. Chinese.
-
Protein arginine methylation: a prominent modification and its demethylation.Cell Mol Life Sci. 2017 Sep;74(18):3305-3315. doi: 10.1007/s00018-017-2515-z. Epub 2017 Mar 31. Cell Mol Life Sci. 2017. PMID: 28364192 Free PMC article. Review.
-
Non-Histone Arginine Methylation by Protein Arginine Methyltransferases.Curr Protein Pept Sci. 2020;21(7):699-712. doi: 10.2174/1389203721666200507091952. Curr Protein Pept Sci. 2020. PMID: 32379587 Free PMC article. Review.
-
Posttranslational modifications of the histone 3 tail and their impact on the activity of histone lysine demethylases in vitro.PLoS One. 2013 Jul 2;8(7):e67653. doi: 10.1371/journal.pone.0067653. Print 2013. PLoS One. 2013. PMID: 23844048 Free PMC article.
-
The emerging therapeutic potential of histone methyltransferase and demethylase inhibitors.ChemMedChem. 2009 Oct;4(10):1568-82. doi: 10.1002/cmdc.200900301. ChemMedChem. 2009. PMID: 19739196 Review.
Cited by
-
Strategies for Cancer Treatment Based on Photonic Nanomedicine.Materials (Basel). 2021 Mar 16;14(6):1435. doi: 10.3390/ma14061435. Materials (Basel). 2021. PMID: 33809479 Free PMC article. Review.
-
(S)-2-(Cyclobutylamino)-N-(3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl)isonicotinamide Attenuates RANKL-Induced Osteoclast Differentiation by Inhibiting NF-κB Nuclear Translocation.Int J Mol Sci. 2023 Feb 21;24(5):4327. doi: 10.3390/ijms24054327. Int J Mol Sci. 2023. PMID: 36901758 Free PMC article.
-
An Updated Review on the Significance of DNA and Protein Methyltransferases and De-methylases in Human Diseases: From Molecular Mechanism to Novel Therapeutic Approaches.Curr Med Chem. 2024;31(23):3550-3587. doi: 10.2174/0929867330666230607124803. Curr Med Chem. 2024. PMID: 37287285 Review.
-
Epigenetics of methylation modifications in diabetic cardiomyopathy.Front Endocrinol (Lausanne). 2023 Mar 15;14:1119765. doi: 10.3389/fendo.2023.1119765. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37008904 Free PMC article. Review.
-
The multi-functionality of UHRF1: epigenome maintenance and preservation of genome integrity.Nucleic Acids Res. 2021 Jun 21;49(11):6053-6068. doi: 10.1093/nar/gkab293. Nucleic Acids Res. 2021. PMID: 33939809 Free PMC article. Review.
References
-
- Boisvert FM, Chénard CA, Richard S. Protein interfaces in signaling regulated by arginine methylation. Sci STKE. 2005;2005:re2. - PubMed

